graphblas-algorithms is a collection of GraphBLAS algorithms written using python-graphblas. It may be used directly or as an experimental backend to NetworkX.
Why use GraphBLAS Algorithms? Because it is fast, flexible, and familiar by using the NetworkX API.
Are we missing any algorithms that you want? Please let us know!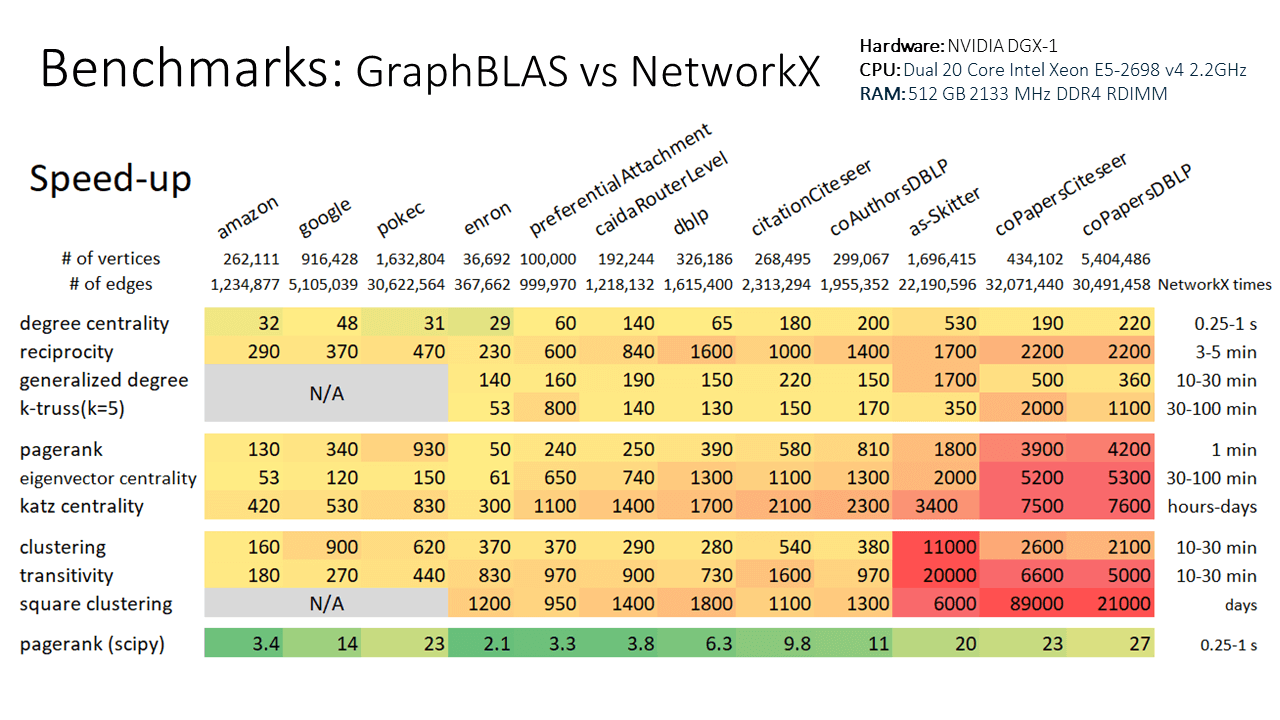
conda install -c conda-forge graphblas-algorithms pip install graphblas-algorithms First, create a GraphBLAS Matrix.
importgraphblasasgbM=gb.Matrix.from_coo( [0, 0, 1, 2, 2, 3], [1, 3, 0, 0, 1, 2], [1., 2., 3., 4., 5., 6.], nrows=4, ncols=4, dtype='float32' )Next wrap the Matrix as ga.Graph.
importgraphblas_algorithmsasgaG=ga.Graph(M)Finally call an algorithm.
hubs, authorities=ga.hits(G)When the result is a value per node, a gb.Vector will be returned. In the case of HITS, two Vectors are returned representing the hubs and authorities values.
Algorithms whose result is a subgraph will return ga.Graph.
Dispatching to plugins is a new feature in Networkx 3.0. When both networkx and graphblas-algorithms are installed in an environment, calls to NetworkX algorithms can be dispatched to the equivalent version in graphblas-algorithms.
importnetworkxasnximportgraphblas_algorithmsasga# Generate a random graph (5000 nodes, 1_000_000 edges)G=nx.erdos_renyi_graph(5000, 0.08) # Explicitly convert to ga.GraphG2=ga.Graph.from_networkx(G) # Pass G2 to NetworkX's k_trussT5=nx.k_truss(G2, 5)G2 is not a nx.Graph, but it does have an attribute __networkx_plugin__ = "graphblas". This tells NetworkX to dispatch the k_truss call to graphblas-algorithms. This link connection exists because graphblas-algorithms registers itself as a "networkx.plugin" entry point.
The result T5 is a ga.Graph representing the 5-truss structure of the original graph. To convert to a NetworkX Graph, use:
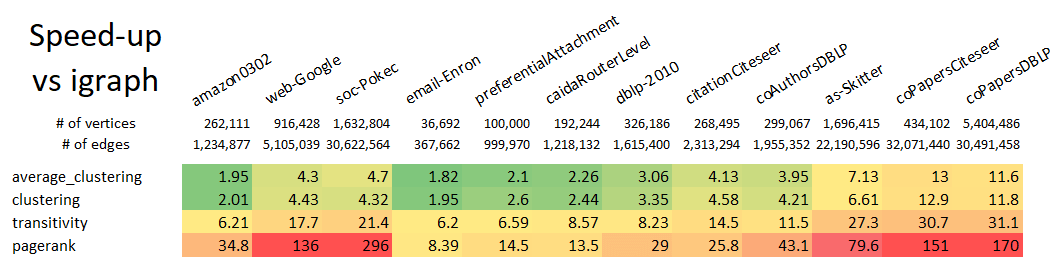
T5.to_networkx()Note that even with the conversions to and from ga.Graph, this example still runs 10x faster than using the native NetworkX k-truss implementation. Speed improvements scale with graph size, so larger graphs will see an even larger speed-up relative to NetworkX.
The following NetworkX algorithms have been implemented by graphblas-algorithms and can be used following the dispatch pattern shown above.
graphblas_algorithms.nxapi ├── boundary │ ├── edge_boundary │ └── node_boundary ├── centrality │ ├── degree_alg │ │ ├── degree_centrality │ │ ├── in_degree_centrality │ │ └── out_degree_centrality │ ├── eigenvector │ │ └── eigenvector_centrality │ └── katz │ └── katz_centrality ├── cluster │ ├── average_clustering │ ├── clustering │ ├── generalized_degree │ ├── square_clustering │ ├── transitivity │ └── triangles ├── community │ └── quality │ ├── inter_community_edges │ └── intra_community_edges ├── components │ ├── connected │ │ ├── is_connected │ │ └── node_connected_component │ └── weakly_connected │ └── is_weakly_connected ├── core │ └── k_truss ├── cuts │ ├── boundary_expansion │ ├── conductance │ ├── cut_size │ ├── edge_expansion │ ├── mixing_expansion │ ├── node_expansion │ ├── normalized_cut_size │ └── volume ├── dag │ ├── ancestors │ └── descendants ├── dominating │ └── is_dominating_set ├── efficiency_measures │ └── efficiency ├── generators │ └── ego │ └── ego_graph ├── isolate │ ├── is_isolate │ ├── isolates │ └── number_of_isolates ├── isomorphism │ └── isomorph │ ├── fast_could_be_isomorphic │ └── faster_could_be_isomorphic ├── linalg │ ├── bethehessianmatrix │ │ └── bethe_hessian_matrix │ ├── graphmatrix │ │ └── adjacency_matrix │ ├── laplacianmatrix │ │ ├── laplacian_matrix │ │ └── normalized_laplacian_matrix │ └── modularitymatrix │ ├── directed_modularity_matrix │ └── modularity_matrix ├── link_analysis │ ├── hits_alg │ │ └── hits │ └── pagerank_alg │ ├── google_matrix │ └── pagerank ├── lowest_common_ancestors │ └── lowest_common_ancestor ├── operators │ ├── binary │ │ ├── compose │ │ ├── difference │ │ ├── disjoint_union │ │ ├── full_join │ │ ├── intersection │ │ ├── symmetric_difference │ │ └── union │ └── unary │ ├── complement │ └── reverse ├── reciprocity │ ├── overall_reciprocity │ └── reciprocity ├── regular │ ├── is_k_regular │ └── is_regular ├── shortest_paths │ ├── dense │ │ ├── floyd_warshall │ │ ├── floyd_warshall_numpy │ │ └── floyd_warshall_predecessor_and_distance │ ├── generic │ │ └── has_path │ ├── unweighted │ │ ├── all_pairs_shortest_path_length │ │ ├── single_source_shortest_path_length │ │ └── single_target_shortest_path_length │ └── weighted │ ├── all_pairs_bellman_ford_path_length │ ├── bellman_ford_path │ ├── bellman_ford_path_length │ ├── negative_edge_cycle │ └── single_source_bellman_ford_path_length ├── simple_paths │ └── is_simple_path ├── smetric │ └── s_metric ├── structuralholes │ └── mutual_weight ├── tournament │ ├── is_tournament │ ├── score_sequence │ └── tournament_matrix ├── traversal │ └── breadth_first_search │ ├── bfs_layers │ └── descendants_at_distance └── triads └── is_triad 




