A novel SARS-CoV-2 related coronavirus in bats from Cambodia
- PMID: 34753934
- PMCID: PMC8578604
- DOI: 10.1038/s41467-021-26809-4
A novel SARS-CoV-2 related coronavirus in bats from Cambodia
Abstract
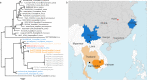
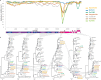
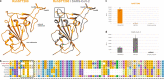
Knowledge of the origin and reservoir of the coronavirus responsible for the ongoing COVID-19 pandemic is still fragmentary. To date, the closest relatives to SARS-CoV-2 have been detected in Rhinolophus bats sampled in the Yunnan province, China. Here we describe the identification of SARS-CoV-2 related coronaviruses in two Rhinolophus shameli bats sampled in Cambodia in 2010. Metagenomic sequencing identifies nearly identical viruses sharing 92.6% nucleotide identity with SARS-CoV-2. Most genomic regions are closely related to SARS-CoV-2, with the exception of a region of the spike, which is not compatible with human ACE2-mediated entry. The discovery of these viruses in a bat species not found in China indicates that SARS-CoV-2 related viruses have a much wider geographic distribution than previously reported, and suggests that Southeast Asia represents a key area to consider for future surveillance for coronaviruses.
© 2021. The Author(s).
Conflict of interest statement
P.B. is currently an employee of GSK vaccines. The remaining authors declare no competing interests.
Figures
Similar articles
- Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses.Cell. 2021 Aug 19;184(17):4380-4391.e14. doi: 10.1016/j.cell.2021.06.008. Epub 2021 Jun 9.Cell. 2021.PMID: 34147139Free PMC article.
- Inferring the ecological niche of bat viruses closely related to SARS-CoV-2 using phylogeographic analyses of Rhinolophus species.Sci Rep. 2021 Jul 12;11(1):14276. doi: 10.1038/s41598-021-93738-z.Sci Rep. 2021.PMID: 34253798Free PMC article.
- Longitudinal monitoring in Cambodia suggests higher circulation of alpha and betacoronaviruses in juvenile and immature bats of three species.Sci Rep. 2021 Dec 17;11(1):24145. doi: 10.1038/s41598-021-03169-z.Sci Rep. 2021.PMID: 34921180Free PMC article.
- [Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70.Vopr Virusol. 2020.PMID: 32515561Review.Russian.
- Overview of Bat and Wildlife Coronavirus Surveillance in Africa: A Framework for Global Investigations.Viruses. 2021 May 18;13(5):936. doi: 10.3390/v13050936.Viruses. 2021.PMID: 34070175Free PMC article.Review.
Cited by
- A Critical Analysis of the Evidence for the SARS-CoV-2 Origin Hypotheses.mSphere. 2023 Apr 20;8(2):e0011923. doi: 10.1128/msphere.00119-23. Epub 2023 Mar 28.mSphere. 2023.PMID: 36897078Free PMC article.
- ACE2 receptor polymorphism in humans and animals increases the risk of the emergence of SARS-CoV-2 variants during repeated intra- and inter-species host-switching of the virus.Front Microbiol. 2023 Jul 13;14:1199561. doi: 10.3389/fmicb.2023.1199561. eCollection 2023.Front Microbiol. 2023.PMID: 37520374Free PMC article.
- The Emergence of the Spike Furin Cleavage Site in SARS-CoV-2.Mol Biol Evol. 2022 Jan 7;39(1):msab327. doi: 10.1093/molbev/msab327.Mol Biol Evol. 2022.PMID: 34788836Free PMC article.Review.
- Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7.Cell Rep. 2021 Jun 29;35(13):109292. doi: 10.1016/j.celrep.2021.109292. Epub 2021 Jun 8.Cell Rep. 2021.PMID: 34166617Free PMC article.
- Material strategies and considerations for serologic testing of global infectious diseases.MRS Bull. 2021;46(9):854-858. doi: 10.1557/s43577-021-00167-4. Epub 2021 Sep 13.MRS Bull. 2021.PMID: 34539056Free PMC article.Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

