A review of studies on animal reservoirs of the SARS coronavirus
- PMID: 17451830
- PMCID: PMC7114516
- DOI: 10.1016/j.virusres.2007.03.012
A review of studies on animal reservoirs of the SARS coronavirus
Abstract
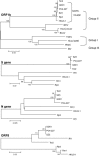
In this review, we summarize the researches on animal reservoirs of the SARS coronavirus (SARS-CoV). Masked palm civets were suspected as the origin of the SARS outbreak in 2003 and was confirmed as the direct origin of SARS cases with mild symptom in 2004. Sequence analysis of the SARS-CoV-like virus in masked palm civets indicated that they were highly homologous to human SARS-CoV with nt identity over 99.6%, indicating the virus has not been circulating in the population of masked palm civets for a very long time. Alignment of 10 complete viral genome sequences from masked palm civets with those of human SARS-CoVs revealed 26 conserved single-nucleotide variations (SNVs) in the viruses from masked palm civets. These conserved SNVs were gradually lost from the genomes of viruses isolated from the early phase to late phase human patients of the 2003 SARS epidemic. In 2005, horseshoe bats were identified as the natural reservoir of a group of coronaviruses that are distantly related to SARS-CoV. The genome sequences of bat SARS-like coronavirus had about 88-92% nt identity with that of the SARS-CoV. The prevalence of antibodies and viral RNA in different bat species and the characteristics of the bat SARS-like coronavirus were elucidated. Apart from masked palm civets and bats, 29 other animal species had been tested for the SARS-CoV, and the results are summarized in this paper.
Figures

Similar articles
- Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12.J Virol. 2015.PMID: 26269185Free PMC article.
- Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4.Infect Genet Evol. 2019.PMID: 30844511Free PMC article.Review.
- Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor.Nature. 2013 Nov 28;503(7477):535-8. doi: 10.1038/nature12711. Epub 2013 Oct 30.Nature. 2013.PMID: 24172901Free PMC article.
- Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms.J Virol. 2005 Sep;79(18):11892-900. doi: 10.1128/JVI.79.18.11892-11900.2005.J Virol. 2005.PMID: 16140765Free PMC article.
- Bats, civets and the emergence of SARS.Curr Top Microbiol Immunol. 2007;315:325-44. doi: 10.1007/978-3-540-70962-6_13.Curr Top Microbiol Immunol. 2007.PMID: 17848070Free PMC article.Review.
Cited by
- Pandemics and food systems - towards a proactive food safety approach to disease prevention & management.Food Secur. 2020;12(4):749-756. doi: 10.1007/s12571-020-01074-3. Epub 2020 Jul 10.Food Secur. 2020.PMID: 32837645Free PMC article.
- Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.Natl Sci Rev. 2022 Jun 23;9(9):nwac122. doi: 10.1093/nsr/nwac122. eCollection 2022 Sep.Natl Sci Rev. 2022.PMID: 36187898Free PMC article.
- Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.J Virol. 2024 Mar 19;98(3):e0115723. doi: 10.1128/jvi.01157-23. Epub 2024 Feb 2.J Virol. 2024.PMID: 38305152Free PMC article.
- Design and validation of consensus-degenerate hybrid oligonucleotide primers for broad and sensitive detection of corona- and toroviruses.J Virol Methods. 2011 Nov;177(2):174-83. doi: 10.1016/j.jviromet.2011.08.005. Epub 2011 Aug 11.J Virol Methods. 2011.PMID: 21864579Free PMC article.
- Exceptional diversity and selection pressure on coronavirus host receptors in bats compared to other mammals.Proc Biol Sci. 2022 Jul 27;289(1979):20220193. doi: 10.1098/rspb.2022.0193. Epub 2022 Jul 27.Proc Biol Sci. 2022.PMID: 35892217Free PMC article.
References
- Cavanagh D., Mawditt K., Adzhar A., Gough R.E., Picault J.P., Naylor C.J., Haydon D., Shaw K., Britton P. Does IBV change slowly despite the capacity of the spike protein to vary greatly? Adv. Exp. Med. Biol. 1998;440:729–734. - PubMed
- Centers for Disease Control and Prevention (CDC), 2003. Prevalence of IgG Antibody to SARS-Associated Coronavirus in Animal Traders—Guangdong Province, China, 2003. Morbidity and Mortality Weekly Report 52, 986–987. - PubMed
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

