Abstract
Objective
Significant morbidity and mortality have occurred in the USA, Europe, and Asia due to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), whereas the numbers of infections and deaths in sub-Saharan Africa (SSA) have remained comparatively low. It has been hypothesized that exposure of the population in SSA to other coronaviruses prior to the COVID-19 pandemic resulted in some degree of cross-protection against SARS-CoV-2 infection and pathogenesis. We evaluated this hypothesis by comparing SARS-CoV-2 cross-reactive antibodies in pre-pandemic plasma samples collected from SSA and the USA.
Method
Pre-COVID-19 pandemic plasma samples from SSA and the USA were collected and tested by immunofluorescence assay against the spike and nucleocapsid proteins of all known human coronaviruses (HCoVs).
Results
The prevalence of SARS-CoV-2 serological cross-reactivity was significantly higher in samples from SSA compared with the USA. Most of these cross-reactive samples cross-recognized the SARS-CoV-2 nucleocapsid protein and the spike proteins of other HCoVs. Nucleocapsid proteins from HCoV-NL63 and HCoV-229E were detected in most samples, thereby implicating prior exposure to these two HCoVs as the likely source of cross-reactive antibodies against SARS-CoV-2.
Conclusion
The low incidences of SARS-CoV-2 infection and disease in SSA appear to be correlated with the pre-pandemic serological cross-recognition of HCoVs, which are substantially more prevalent in SSA than the USA.
Keywords: SARS-CoV-2, COVID-19, Cross-reactivity, Sub-Saharan Africa, Serology, Human coronavirus, HCoV-NL63, HCoV-229E
Introduction
The first case of the COVID-19 pandemic was reported in late 2019 in Wuhan, China, and its causative agent the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) subsequently spread rapidly worldwide (Lu et al., 2020). SARS-CoV-2 is a betacoronavirus and a close relative of the original SARS and Middle East respiratory syndrome coronavirus (MERS) which both cause lethal diseases in human (Chen et al., 2020, Gussow et al., 2020). Four other less pathogenic human coronaviruses (HCoVs) comprising HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E cause mild upper respiratory tract diseases referred to as the “common cold” (Chen et al., 2020, Gussow et al., 2020).
At the time of writing, the COVID-19 pandemic has resulted in over 31 million confirmed SARS-CoV-2 infections and nearly a million deaths, with about 22% of the confirmed cases and 21% of the confirmed deaths occurring in the USA (Nuzzo et al., 2020). Several factors support the hypothesis that populations in sub-Saharan Africa (SSA) might be more susceptible to coronaviral infection and disease, including the high infectious disease burden (Ebola, yellow fever, and cholera outbreaks, as well as high endemic prevalence of HIV-1, tuberculosis, malaria, and parasitic diseases), numerous socioeconomic factors, poor hygiene, nutritional deficiency, and lack of health care access in rural areas (Oleribe et al., 2015, Semeere et al., 2016). The infrastructure for diagnostics and epidemiological surveillance is suboptimal in Africa, but the COVID-19 case mortality rates are lower where large scale testing has been possible compared with elsewhere in the world. There have been no reports from SSA of any abnormal increases in the numbers of respiratory diseases or deaths, which are the hallmarks of the COVID-19 pandemic. Despite the high number of COVID-19 cases and mortality in the USA, Europe, and Asia, the COVID-19 disease burden has remained surprisingly low in SSA (Nuzzo et al., 2020). A potential explanatory factor could be the relatively younger African populations compared with those in the USA or Europe, which may have resulted in more asymptomatic cases (Gaye et al., 2020). In addition, the onerous high infectious disease burden in SSA may have included exposure to other HCoVs, which could have elicited humoral responses against conserved epitopes among coronaviruses to engender cross-protection. This prior exposure to other coronaviruses may offer some level of cross-protective immune response against SARS-CoV-2 infection, thereby reducing the number and/or severity of COVID-19 cases.
To investigate this hypothesis, we examined pre-COVID-19 pandemic plasma samples from Tanzania, Zambia, and the USA to determine their serological cross-reactivity against the spike and nucleocapsid proteins of SARS-CoV-2 and other HCoVs (SARS, MERS, HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E), as well as whether HIV-1 infection, which is endemic in SSA, might affect the prevalence of serological cross-reactive against SARS-CoV-2. We found that pre-COVID-19 pandemic SSA samples had a significantly higher prevalence of serological cross-reactivity against SARS-CoV-2 than samples from the USA. In addition, the SARS-CoV-2 cross-reactive plasma samples strongly recognized the spike and nucleocapsid proteins from specific human seasonal coronaviruses, thereby suggesting that prior exposure to these other coronaviruses may have induced partially protective responses against SAR-CoV-2.
Materials and methods
Study cohort and samples
The study cohort comprised 289 consenting subjects aged ≥18 years and belonging to both genders from Dar es Salaam, Tanzania; Lusaka, Zambia; and Lincoln, Nebraska, USA. The Tanzanian samples comprised 105 plasma samples collected from voluntary blood donors between March and May, 2019. The Zambian samples comprised 99 plasma samples collected between 2017 and early 2019. The plasma samples from the USA were collected from 85 blood donors during 2005, 2007, and 2009 in Lincoln, Nebraska, and they were also evaluated for comparison. All study procedures were approved by the Institutional Review Boards at the Tanzania National Institute for Medical Research, Ocean Road Cancer Institute, University of Zambia Biomedical Research Ethics Committee, and the University of Nebraska-Lincoln.
HIV serological testing
HIV-1 serology testing was conducted using the HIV Rapid Test Algorithm (United Republic of Tanzania, 2007) in Tanzania and the Alere Determine HIV-1/2 Ag/Ab Combo test in Zambia (Abbott Laboratories, Chicago, IL, USA). The serological results were verified in our laboratory at Lincoln, Nebraska using the HIV-1-2.0 First Response kit (Premier Medical Corporation Limited, Daman, India).
Immunofluorescence assay (IFA) against SARS-CoV-2 and other HCoVs
To detect the presence of serological cross-reactivity against SARS-CoV-2 and other HCoVs, we used an IFA against the spike and nucleocapsid proteins of SARS, SARS-CoV-2, MERS, HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E. Briefly, HEK-293T cells (American Type Culture Collection [ATCC], Manassas, VA, USA) were transfected with mammalian expression plasmids encoding either the spike or nucleocapsid proteins of the respective coronaviruses (Addgene, Watertown, MA, USA and Sino Biological, Wayne, PA, USA). After 48 h, the transfected cells were fixed and seeded onto 12-well polytetrafluoroethylene printed slides (Electron Microscopy Sciences, Hatfield, PA, USA), where each well contained either spike, nucleocapsid, or mock transfected cells, before permeabilization with 0.3% hydrogen peroxide (H2O2) methanol solution. The prepared IFA slides were stored at −80 °C.
Plasma samples were diluted 1:20 with phosphate-buffered saline (PBS) plus 0.1% Tween-20 and incubated at room temperature for 30 min. The prepared IFA slides were thawed and incubated with PBS plus 0.1% Tween-20 for 30 min at 37 °C. Each diluted plasma sample was then added to cells expressing each HCoV antigen or control wells and incubated for 1 h at 37 °C. After primary antibody binding and washing, a secondary mouse monoclonal anti-human immunoglobulin G (IgG) antibody (ATCC, USA) was bound for 1 h at 37 °C, before washing to remove any excess unbound antibody. Tertiary CY2-conjugated donkey anti-mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA, USA) was then added and incubated for 1 h at 37 °C. Finally, the slides were counterstained with 0.004% Evans blue solution for 30 s. All IFA slides were washed 3 times with PBS after each incubation step. The stained IFA slides were read by three independent readers using a Nikon Eclipse 50i fluorescence microscope. Positive cells were enumerated based on green fluorescence against a red cellular counterstain. A well was only considered positive or negative if at least two independent readers were concordant in reporting the outcome. Summarization of results and statistical analysis (two-tailed Fisher’s exact test) were conducted and plotted using GraphPad (GraphPad Software, San Diego, CA, USA).
Results
To evaluate the serological cross-reactivity against SARS-CoV-2 and other HCoVs, we obtained blood donor plasma samples from Tanzania (n = 105), Zambia (n = 99), and the USA (n = 85) (Table 1 ). These samples were collected between 2005 and May 2019, and before the current COVID-19 pandemic, but synchronous plasma samples were not available due to the retrospective nature of the study. Among our cohort, 6.7% and 43.4% of the Tanzanian and Zambian samples, respectively, were HIV-1 positive. By contrast, all of the plasma samples collected in the USA were HIV-1 negative. The high prevalence of HIV-1 infection in the Zambian samples does not reflect the national HIV-1 infection rate, but instead it was intended to support comparisons of the cross-reactivity against SARS-CoV-2 and the recognition of other coronaviruses in HIV-1 positive and negative subjects.
Table 1.
Study cohort and sampling time periods.
| Country | Sample size | HIV-1 positive (%) | Sampling time period |
|---|---|---|---|
| Tanzania | 105 | 7 (6.7%) | March to May 2019 |
| Zambia | 99 | 43 (43.4%) | 2017 to early 2019 |
| USA | 85 | 0 (0%) | 2005, 2007, and 2009 |
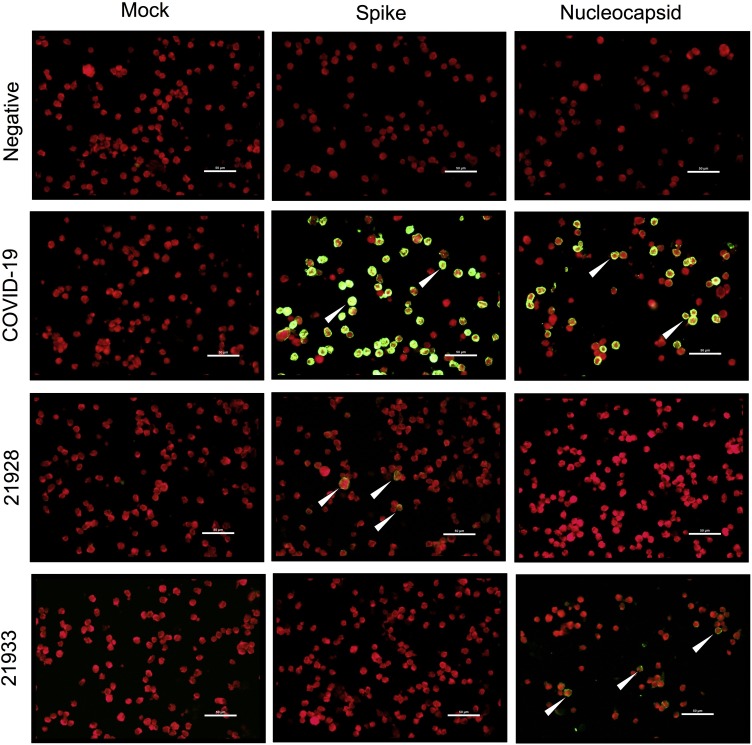
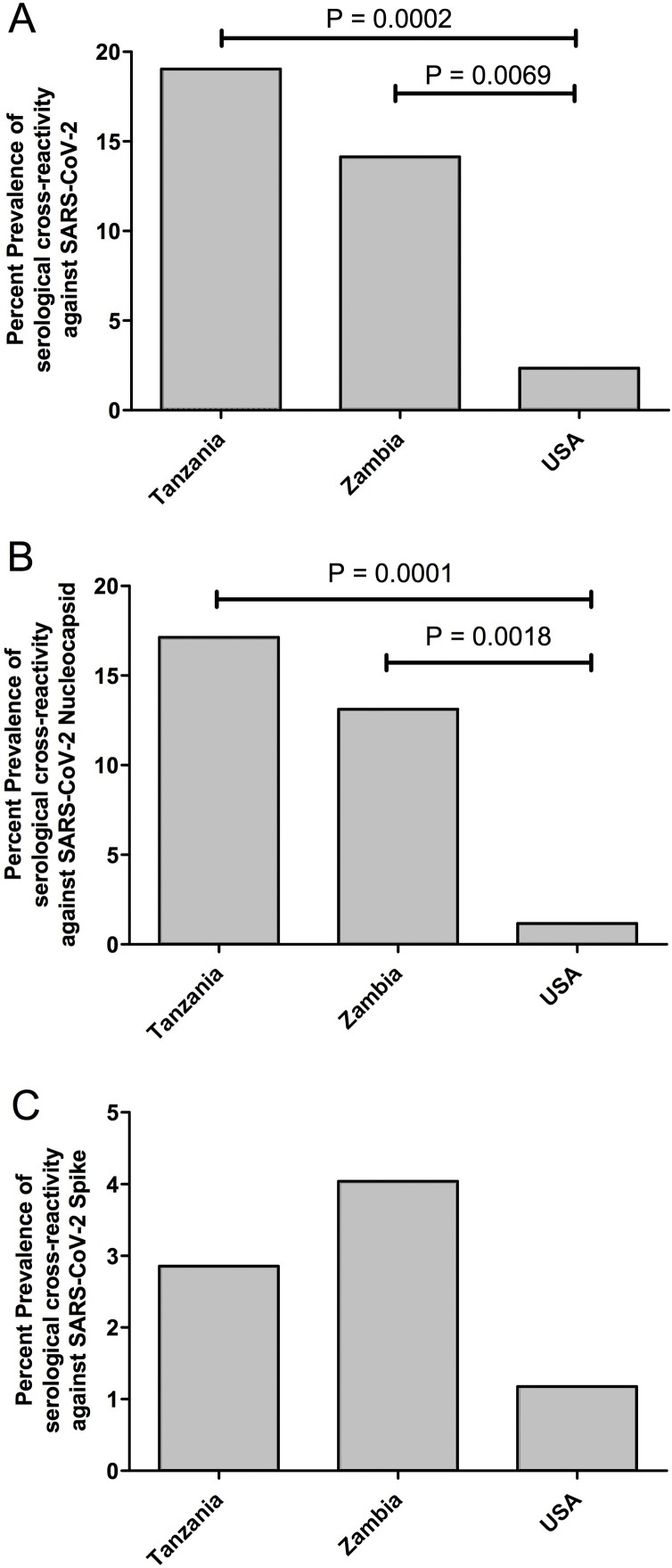
The plasma samples were screened for cross-reactivity against SARS-CoV-2 using an IFA. As shown in Figure 1 , COVID-19 convalescent positive control plasma resulted in strong green fluorescence staining in cells expressing either the SARS-CoV-2 spike or nucleocapsid proteins, but not in the mock transfected cells. Green fluorescence was not evident on antigen-expressing cells stained with negative control plasma, thereby demonstrating the specificity of the IFA for detecting SARS-CoV-2 specific IgG antibodies. Interestingly, green fluorescence was evident on cells expressing either the SARS-CoV-2 spike or nucleocapsid proteins when stained with some pre-COVID-19 pandemic plasma samples, which indicates the presence of antibodies cross-reactive against SARS-CoV-2 prior to the current COVID-19 pandemic (Figure 1). Compared with samples from the USA (2.4%), the prevalence of serological cross-reactivity against SARS-CoV-2 was significantly higher in Tanzania (19%) (P = 0.0002) and Zambia (14.1%) (P = 0.0069) (Figure 2 A). A breakdown of the anti-SARS-CoV-2 cross-reactivity indicated that most of the Tanzanian and Zambian cross-reactive responses targeted the SARS-CoV-2 nucleocapsid protein, with 17.1% (P = 0.0001) and 13.1% (P = 0.0018), respectively, and these levels were significantly higher than those in samples from the USA, i.e., 1.2% (Figure 2B). There were no statistical differences in the anti-SARS-CoV-2 spike cross-reactivity prevalence rates, with 2.9% in Tanzania, 4% in Zambia, and 1.2% in the USA (Figure 2C). In addition, none of the cross-reactive samples from Tanzania were HIV-1 positive and only 5/43 (11.6%) HIV-1 positive samples from Zambia were cross-reactive toward SARS-CoV-2. Moreover, 9/56 (16%) HIV-1 negative samples from Zambia were cross-reactive toward SARS-CoV-2. Therefore, HIV-1 infected individuals appeared to have a lower cross-reactive response toward SARS-CoV-2. However, an HIV-1 positive cohort with a larger sample size will be needed to verify this observation.
Figure 1.
IFA against either mock, SARS-CoV-2 spike, or nucleocapsid expressing cells. Representative images are shown for IFA with negative control plasma, COVID-19 convalescence plasma (positive control), and pre-COVID-19 pandemic cross-reactive plasma samples 21928 and 21933. Sample 21928 exhibited cross-reactivity against SARS-CoV-2 spike, but not its respective mock and SARS-CoV-2 nucleocapsid. Sample 21933 exhibited cross-reactivity against SARS-CoV-2 nucleocapsid, but not its respective mock and SARS-CoV-2 spike. White arrows indicate positive cells. Scale bar represent 50 μm.
Figure 2.
Percentage prevalence of serological cross-reactivity against SARS-CoV-2 in Tanzania, Zambia, and the USA. (A) Combined serological cross-reactivity against SARS-CoV-2 spike and nucleocapsid. (B) Serological cross-reactivity against SARS-CoV-2 nucleocapsid. (C) Serological cross-reactivity against SARS-CoV-2 spike.
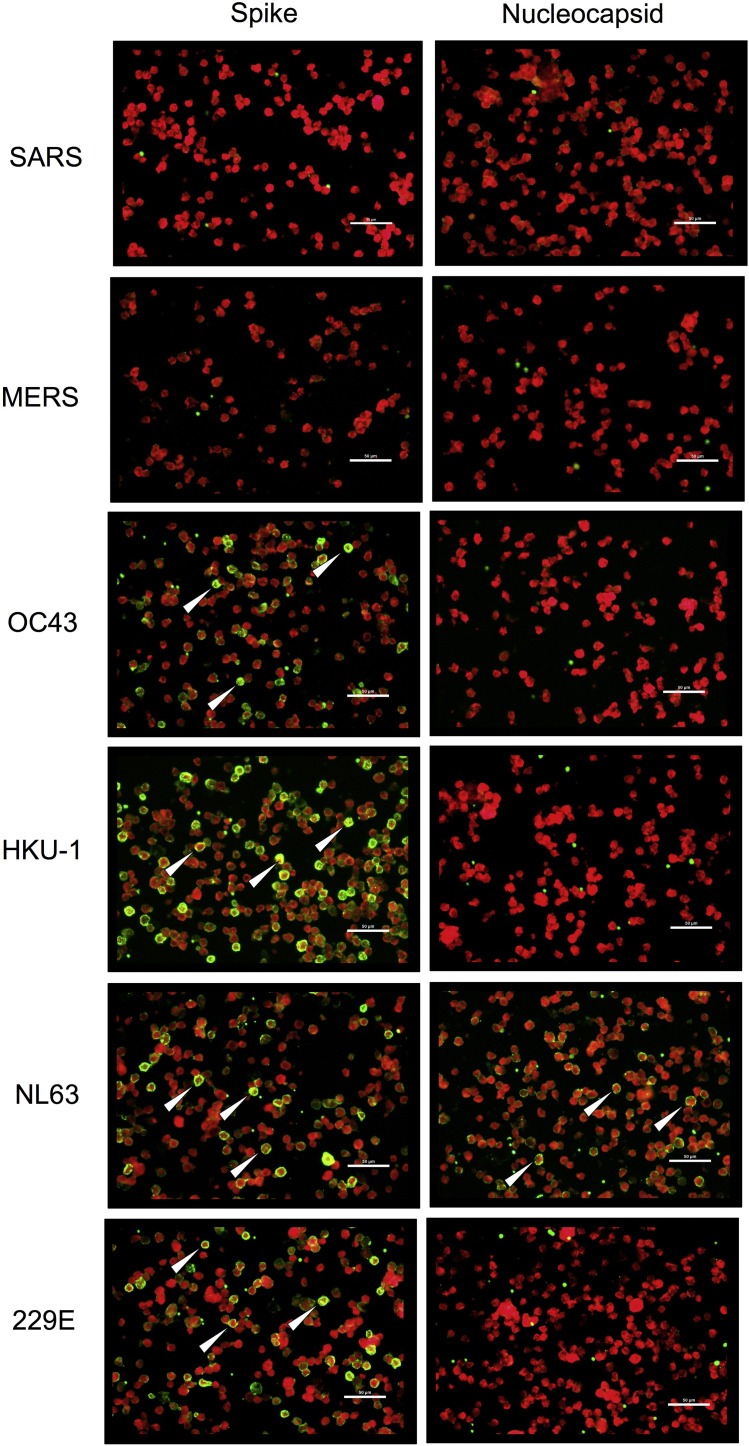
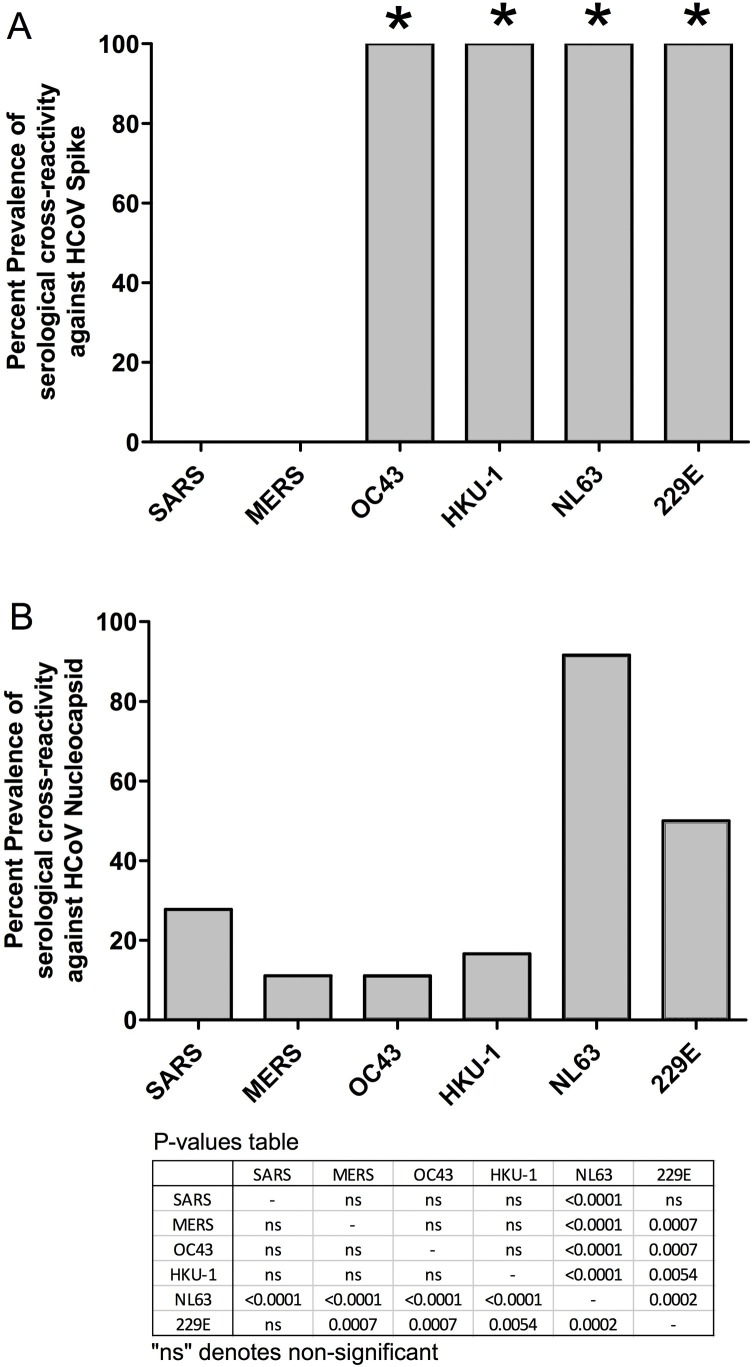
To investigate whether anti-SARS-CoV-2 cross-reactivity correlated with past exposure to other HCoVs, pre-COVID-19 pandemic plasma samples that cross-reacted against SARS-CoV-2 were tested to determine their anti-HCoV responses. As demonstrated by a representative cross-reactive plasma sample (21854), IFA against the spike and nucleocapsid proteins of different HCoVs detected IgG antibodies against the HCoV-OC43, HKU-1, NL63, and 229E spike proteins (Figure 3 ). However, the same plasma sample only recognized the nucleocapsid of HCoV-NL63, which suggests that HCoV-NL63 was the main source of antigenic exposure for this individual. When we analyzed all of the SARS-CoV-2 serologically cross-reactive samples, we found that 100% recognized the spike proteins from all four HCoVs that cause the common cold, but not those from SARS and MERS (Figure 4A). This difference in the recognition of the common HCoV spike proteins versus the SARS and MERS spike proteins was statistically significant (P < 0.0001). In addition, comparisons of HCoV nucleocapsid recognition among all of the samples showed that the most commonly recognized nucleocapsid was that of HCoV-NL63, followed by HCoV-229E, with 92% and 50%, respectively (Figure 4B). This difference was statistically significant compared with the recognition of the other HCoVs, with P-values ranging from <0.0001 to 0.0002 for HCoV-NL63, and P-values ranging from 0.0002 to 0.0054 for HCoV-229E (Figure 4B). Finally, we compared how individuals from different countries responded against various HCoV nucleocapsid proteins. Qualitative analysis showed that the Zambian SARS-CoV-2 cross-reactive samples tended to recognize a wider range of HCoVs compared with the samples from Tanzania (Table 2 ). Some Zambian individuals recognized four to six different HCoVs, whereas Tanzanian individuals recognized a maximum of three different HCoVs. However, the small sample size limited the statistical analysis of this difference.
Figure 3.
IFA against SARS, MERS, HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E spike or nucleocapsid expressing cells. Representative images are shown of IFA with pre-COVID-19 pandemic cross-reactive plasma sample 21854. Sample 21854 strongly recognized the spike proteins of HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E, but not those of SARS and MERS. Sample 21854 only recognized the nucleocapsid of HCoV-NL63 but not those of the other human coronaviruses. White arrows indicate positive cells. Scale bar represent 50 μm.
Figure 4.
Percentage prevalence of serological cross-reactivity against SARS, MERS, HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E. (A) Spike. (B) Nucleocapsid.
Table 2.
Individual cross-reactive responses against the nucleocapsid proteins of SARS, MERS, HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E.
| Country | Sample ID | SARS | MERS | OC43 | HKU-1 | NL63 | 229E |
|---|---|---|---|---|---|---|---|
| Tanzania | 21850 | – | – | – | – | + | – |
| Tanzania | 21854 | – | – | – | – | + | – |
| Tanzania | 21868 | – | – | – | – | + | + |
| Tanzania | 21872 | – | – | – | – | + | + |
| Tanzania | 21873 | – | – | – | – | + | – |
| Tanzania | 21928 | – | – | – | – | + | – |
| Tanzania | 21933 | – | – | – | – | + | – |
| Tanzania | 211141 | + | – | – | – | + | + |
| Tanzania | 211145 | – | – | – | – | + | + |
| Tanzania | 211157 | – | – | – | + | + | + |
| Tanzania | 211176 | + | – | – | – | + | – |
| Tanzania | 211177 | – | – | – | – | + | – |
| Tanzania | 211181 | – | – | – | – | + | + |
| Tanzania | 211182 | – | – | – | – | + | – |
| Tanzania | 211185 | – | – | – | + | + | + |
| Tanzania | 211188 | – | – | – | – | + | + |
| Tanzania | 211192 | – | + | – | – | + | + |
| Tanzania | 211203 | – | – | – | – | + | + |
| Tanzania | 211205 | – | – | – | – | + | + |
| Tanzania | 211210 | – | – | – | – | + | + |
| Zambia | C3076 | + | – | – | – | + | – |
| Zambia | C3082 | – | – | – | – | + | – |
| Zambia | C3154 | – | – | – | – | + | – |
| Zambia | C3155 | + | – | + | + | + | + |
| Zambia | C3156 | – | – | – | – | + | + |
| Zambia | C3163 | + | – | + | – | + | + |
| Zambia | C3166 | – | + | – | – | + | – |
| Zambia | C3182 | + | – | + | + | + | + |
| Zambia | C3187 | – | – | – | – | – | – |
| Zambia | C3197 | – | – | – | – | + | – |
| Zambia | C3202 | + | + | + | + | + | + |
| Zambia | C3204 | + | + | – | – | – | – |
| Zambia | N044 | + | – | – | – | + | – |
| Zambia | N216 | + | – | – | – | + | – |
| USA | KC-34 | – | – | – | – | + | + |
| USA | KC-65 | – | – | – | – | + | + |
HCoV, human coronaviruses; MERS, Middle East respiratory syndrome coronavirus; SARS, severe acute respiratory syndrome coronavirus-2.
Discussion
Despite the rapid spread of SARS-CoV-2, which has caused nearly a million deaths worldwide to date, the SARS-CoV-2 burden remains surprisingly low in SSA. This is particularly surprising given the high prevalence of other diseases such as HIV-1, malaria, cancer, and tuberculosis, as well as inadequate health care and the impact of poverty. The current SARS-CoV-2 disease burden is much higher in the USA than countries in SSA. It is not known whether the low prevalence of serological cross-reactivity to HCoV in the USA, as shown in the present study, is directly associated with the outcomes of the COVID-19 pandemic in the USA. Our data suggest that populations in SSA were previously exposed to a spectrum of HCoVs, which provided some cross-reactivity against SARS-CoV-2 and this may have limited the number of infections or pathogenesis in SSA. In support of this hypothesis, we detected serological cross-reactivity against SARS-CoV-2 antigens in pre-COVID-19 plasma samples from Tanzania and Zambia at levels nearly 8 and 6 times higher, respectively, than the prevalence in samples from the USA. By comparing the prevalence of serological cross-reactivity against SARS-CoV-2 between HIV-1 positive and negative Zambian individuals, we also found that HIV-1 infection appeared to lower the cross-reactive response toward SARS-CoV-2, which could have been due to a weakened immune response in HIV-1 infected individuals. However, an HIV-1 positive cohort with a larger sample size will be needed to confirm this observation.
Individuals in our study cohort who reacted to SARS-CoV-2 antigens predominantly cross-reacted with the SARS-CoV-2 nucleocapsid protein. Consistent with spike protein variation across coronaviruses, few individuals reacted with the SARS-CoV-2 spike protein. This also supported the indication that the SARS-CoV-2 spike protein is a more specific target for serological testing of SARS-CoV-2 infection and the humoral response to infection. Conversely, a recent study suggested that the SARS-CoV-2 nucleocapsid is more sensitive than the spike protein for the early detection of SARS-CoV-2 infection (Burbelo et al., 2020), thereby highlighting the distinction between sensitivity and specificity. The results obtained based on our analysis of pre-COVID-19 pandemic samples support the hypothesis that detecting SARS-CoV-2 infection with the nucleocapsid may generate a significant number of false positive results, which could be country specific, with countries such as Tanzania and Zambia potentially having higher false positive rates than the USA due to prior exposure to other coronaviruses.
We also investigated which HCoV was responsible for the observed cross-reactivity with SARS-CoV-2 and found that all SARS-CoV-2 cross-reactive samples strongly cross-reacted with the spike proteins from HCoV-OC43, HCoV-HKU-1, HCoV-NL63, and HCoV-229E, but not those from SARS or MERS. These findings suggest that some immunogenic epitopes within the spike protein may be shared among all known HCoVs. In addition, most of our SARS-CoV-2 cross-reactive samples reacted strongly against the nucleocapsid proteins of HCoV-NL63 and HCoV-229E, thereby suggesting that these two HCoVs may have served as the source of antigenic exposure in SSA prior to the COVID-19 pandemic. Cross-reactivity against SARS-CoV-1 nucleocapsid as a result of exposure to HCoVs, such as HCoV-OC43, has been reported previously (Patrick et al., 2006), but the present study is the first to link HCoV-NL63 and HCoV-229E to cross-reactivity against SARS-CoV-2 in the SSA.
HCoV-NL63 and HCoV-229E are alphacoronaviruses, whereas HCoV-OC43 and HCoV-HKU-1 are betacoronaviruses like SARS-CoV-2 (Abdul-Rasool and Fielding, 2010). In addition, HCoV-NL63 is the only other HCoV that uses angiotensin-converting enzyme 2, which is the same receptor used by SARS and SARS-CoV-2 (Abdul-Rasool and Fielding, 2010, Hofmann et al., 2005). The epidemiology of HCoV-NL63 and HCoV-229E is poorly defined in adults. Some studies reported a prevalence rate of 8.8% for HCoV-NL63 in the USA but less than 1% in the UK (Esper et al., 2005, Gaunt et al., 2010). The prevalence of HCoV-229E is unclear. Importantly, no epidemiological data exist for these two HCoVs in SSA. A recent bioinformatics study also suggested that SARS-CoV-2 evolved from a bat coronavirus and bats may be a primary reservoir (Boni et al., 2020). Given the abundant wildlife in Africa, including multiple species of bats, and their often close proximity to humans, we cannot exclude the possibility of exposure to zoonotic coronaviruses eliciting the observed cross-reactivity against SARS-CoV-2 and other HCoVs. Our results suggest that infections with HCoV-NL63 or similar transmissible zoonotic agents were common in SSA prior to the COVID-19 pandemic.
Finally, the functions of these SARS-CoV-2 cross-reactive antibodies and whether they provide any protection against SARS-CoV-2 infection or disease progression is still unclear, and these questions cannot be resolved with retrospective cross-sectional sampling. The SARS-CoV-2 nucleocapsid is the major antigen recognized by these cross-reactive antibodies, so we suggest that antibody-dependent effector mechanisms such as antibody-dependent cellular cytotoxicity could play protective roles. Our detection of SARS-CoV-2 cross-reactive antibodies in pre-COVID-19 pandemic samples supports the finding obtained in a recent study, which showed that exposure to HCoV/common cold induced SARS-CoV-2 cross-reactive T-cell responses in pre-pandemic samples (Mateus et al., 2020). Both adaptive responses may have offered some protection against COVID-19 pathogenesis if not SARS-CoV-2 infection. A limitation of our study is that no peripheral blood mononuclear cells were collected prior to the COVID-19 pandemic in order to analyze the potential cross-reactive T-cell response. Thus, a larger sample size and more in-depth longitudinal analysis of the functions of these cross-reactive antibodies, as well as the cross-reactive T-cell response will be needed in future studies.
Conflict of interest
All authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author contributions
FYT, SJL, PBP, and AAC performed IFA. PBP and AAC performed HIV serological testing. ON, PJ, JRN, JM, and JTW collected all plasma samples. FYT wrote the manuscript. CW supervised all aspects of the study. All authors reviewed and approved the manuscript.
Acknowledgements
This study was supported partly by the US National Institute of Health (NIH) - National Cancer Institute U54 CA190155 (CW) and U54 CA221204 (CW); and Fogarty International Center K43TW011418 (SJL), K43TW011095 (ON), and D43 TW010354 (CW). ON is a Fogarty fellow and PBP is an INBRE scholar supported by National Institute of General Medical Sciences P20 GM103427.
References
- Abdul-Rasool S., Fielding B.C. Understanding human coronavirus HCoV-NL63. Open Virol J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Tian E.K., He B., Tian L., Han R., Wang S. Overview of lethal human coronaviruses. Signal Transduct Target Ther. 2020;5(1):89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005;191(4):492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaye B., Khoury S., Cene C.W., Kingue S., N’Guetta R., Lassale C. Socio-demographic and epidemiological consideration of Africa’s COVID-19 response: what is the possible pandemic course? Nat Med. 2020;26(7):996–999. doi: 10.1038/s41591-020-0960-y. [DOI] [PubMed] [Google Scholar]
- Gussow A.B., Auslander N., Faure G., Wolf Y.I., Zhang F., Koonin E.V. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci U S A. 2020;117(26):15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzzo J., Moss B., Kahn J., Rutkow L., Laboratory A.P., Gardner L. 2020. Johns Hopkins Coronavirus Resource Center.https://coronavirus.jhu.edu/ Available from: [Google Scholar]
- Oleribe O.O., Salako B.L., Ka M.M., Akpalu A., McConnochie M., Foster M. Ebola virus disease epidemic in West Africa: lessons learned and issues arising from West African countries. Clin Med (Lond) 2015;15(1):54–57. doi: 10.7861/clinmedicine.15-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeere A., Wenger M., Busakhala N., Buziba N., Bwana M., Muyindike W. A prospective ascertainment of cancer incidence in sub-Saharan Africa: the case of Kaposi sarcoma. Cancer Med-Us. 2016;5(5):914–928. doi: 10.1002/cam4.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Republic of Tanzania MoHaSWN. 2007. Guidelines on HIV Testing and Counseling in Clinical Settings.http://www.who.int/hiv/topics/vct/TZ_PITC-Guidelines Available from: [Google Scholar]