Abstract
INTRODUCTION:
The 2019 coronavirus disease (COVID-19) is a major global health concern. Joint efforts for effective surveillance of COVID-19 require immediate transmission of reliable data. In this regard, a standardized and interoperable reporting framework is essential in a consistent and timely manner. Thus, this research aimed at to determine data requirements towards interoperability.
MATERIALS AND METHODS:
In this cross-sectional and descriptive study, a combination of literature study and expert consensus approach was used to design COVID-19 Minimum Data Set (MDS). A MDS checklist was extracted and validated. The definitive data elements of the MDS were determined by applying the Delphi technique. Then, the existing messaging and data standard templates (Health Level Seven-Clinical Document Architecture [HL7-CDA] and SNOMED-CT) were used to design the surveillance interoperable framework.
RESULTS:
The proposed MDS was divided into administrative and clinical sections with three and eight data classes and 29 and 40 data fields, respectively. Then, for each data field, structured data values along with SNOMED-CT codes were defined and structured according HL7-CDA standard.
DISCUSSION AND CONCLUSION:
The absence of effective and integrated system for COVID-19 surveillance can delay critical public health measures, leading to increased disease prevalence and mortality. The heterogeneity of reporting templates and lack of uniform data sets hamper the optimal information exchange among multiple systems. Thus, developing a unified and interoperable reporting framework is more effective to prompt reaction to the COVID-19 outbreak.
Keywords: COVID-19, coronavirus disease 2019, minimum data set, semantic interoperability, surveillance system
Introduction
In December 2019, a cluster of pneumonia cases of primary unknown etiology emerged in Wuhan City, Hubei Province, China. After extensive speculation, ultimately, a novel species of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was recognized as the causative pathogen of the disease. The disease name was initially called “2019 novel CoV” and later changed into CoV disease 2019 (COVID-19). The highly contagious nature of the disease and rapid increase of emerging new cases in China and many other countries have led the World Health Organization (WHO) on January 30, 2020, to declare the COVID-19 outbreak a global public health threat.[1,2,3,4,5,6,7,8]
Surveillance is the foundation of public health practice and research. To prepare for and deal with COVID-19 pandemic outbreak, a robust and responsive surveillance system should be considered, which provides a partnership cooperation among public health practitioners, clinicians, and policymakers to direct disease control and prevention efforts.[9,10] The effectiveness of COVID-19 Surveillance System (COVSS) depends on clinical data and reports from wide scattered public and hospital information system as data input (e.g., Hospital information systems (HIS), Iranian Electronic Health Record (so-called SEPAS), Iranian Integrated Health System (known as SIB), and other clinical information systems). In this sense, effective implementation of COVSS necessitates clear and coherent sets of data, along with unified standards for sharing this data rapidly, supporting e-health and P4-medicine (Predictive, Preventive, Personalized, and Participatory).[11,12] A modular methodology should be developed in the design and implementation of information systems that will increase their integrity and enterprise usefulness. Data standardization and harmonization is the first important step in the life cycle of the information system (known as System Development Life Cycle (SDLC)) and it should be achieved conforming to a proper plan.[13,14] Minimum Data Set (MDS) is one standard approach for data collection, providing accurate access to health data. In respect to the development Public Health Surveillance (PHS), MDS solution offers enhanced progresses in systematic collection, interpretation, comparison, and integration of data regarding health-related threats. However, data sharing may also be hindered if standardized methods are not used for coding and formatting data. The use of Information and Communication Technology may aid in enabling standardized, automated, and interoperable frameworks for data exchange between public and health information systems with heterogeneous platforms.[15,16,17,18,19] Thus, the present study was conducted to provide a comprehensive MDS as a template for implementing a COVSS and then presented designing an exchanging framework toward interoperability in the context of COVID-19.
Materials and Methods
This was a cross-sectional descriptive study conducted in 2020. Initially, to design the COVID-19 MDS, a combination of literature review and expert consensus approach was used. In this regard, a review of the literature was conducted to retrieve related data resources on COVID-19, while also applying guidelines and instructions issued from local, national, and international organizations, especially the WHO and Center for Disease Control. Literature review was limited to English languages between December 2019 and March 2020 in the full text along with valid sources available on PubMed, Scopus, Web of Science, Science direct, Embase, and Cochrane databases.
To confirm the COVID-19 MDS, the preliminary data list was evaluated through consensus of the selected experts after review and discussion. Thus, we brought together a multidisciplinary team of 40 samples with expertise in virology, epidemiology, public health practitioners, infectious diseases, and experience in health information management. A researcher-made questionnaire was created to validate data fields. The experts participating in the study were asked to review the initial draft of variables to score the items according to the importance perceived by them based on a 5-point Likert scale (ranging from 1:“very slightly important” to 5:”highly important”.[1,2,3,4,5]
The content validity of the questionnaire was evaluated using the comments from medical informatics and health information technology experts (a total of six persons, consisting of three experts in each field). For the reliability of the questionnaire, the test–retest method was used by 10 infectious disease specialists. Through decision Delphi technique in two rounds, decisions on included data fields were made based on the agreement level. Specifically, data fields with <50% agreement were excluded in the first round, while those with more than 75% agreement were included in the primary round. Those with 50%–75% agreement were surveyed in the second round, and if there was 75% consensus over a subject, it was regarded as a final data field. Further, if any experts intended to change, delete, or add a variable for a specific purpose, they were asked to write an acceptable reason. The collected data were analyzed by SPSS 16 where Spearman's rank correlation coefficient was used to evaluate the reliability of the questionnaire, which showed a coefficient of 85%.
To determine the corresponding information content of data fields, a complete COVID-19 patient record sample in the Ayatollah Taleghani Hospital (focal center of COVID-19, Abadan, Iran) was selected and its contents were extracted by a checklist. Then, the information content was coded using selected classification or nomenclature systems.
In the next step, all scattered codes were mapped to Systematized Nomenclature of Medicine–Clinical Terms (SNOMED-CT) reference codes using NPEX SNOMED-CT online browser (https://snomedbrowser.com/). This process was visualized through MindMaple Lite 1.71 software as a graphic user interface representing thesaurus mapping across multiple medical terminologies [Figure 1]. Finally, SNOMED-CT codes were structured into Health Level Seven-Clinical Document Architecture (HL7-CDA) standard framework to provide the message syntax. Finally, the Extensive Markup Language (XML) hierarchical rules were defined for standardization of the message structure. XML provides a comprehensive and unified human- and machine-readable resource which formally defines and represents CDA information as a set of concepts in a given domain. Overall, the CDA schema was designed based on coded and structured title and body (CDA, level two and three) through SNOMED-CT reference codes and XML structure.
Figure 1.
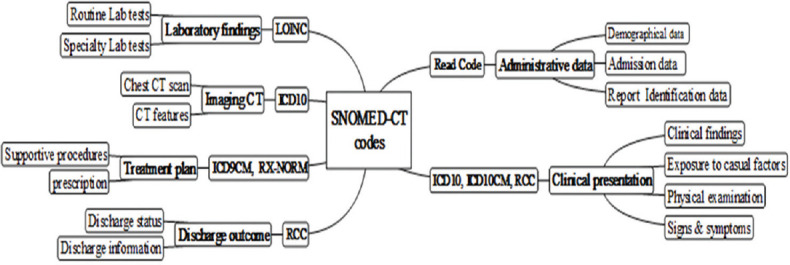
MindMaple Lite1.71 routes
Results
After the literature review, the proposed COVID-19 MDS was divided into administrative and clinical data categories. Each of the categories contained three and eight data class and 52 and 85 data field, respectively. The administrative data category included demographical, admission, and report ID data classes. The second category was clinical data involving clinical presentation, exposure to casual factors, physical examination, signs and symptoms, laboratory findings, CT results, treatment plan, and discharge outcome. Then, Delphi surveys were used to finalize the primary MDS. The results of two Delphi rounds are presented in Table 1.
Table 1.
Administrative and clinical data classes for a minimum data set for coronavirus disease-19 reporting
| Data classes | Total number of fields | First round of Delphi | Second round of Delphi | Final | ||||
|---|---|---|---|---|---|---|---|---|
| <50% | 50%-75% | 75%< | <50% | 50%-75% | 75%< | |||
| Administrative data category | ||||||||
| Demographical | 27 | 6 | 12 | 9 | 6 | 0 | 6 | 15 |
| Admission | 12 | 4 | 3 | 5 | 2 | 0 | 1 | 6 |
| Report ID | 13 | 3 | 5 | 5 | 2 | 0 | 3 | 8 |
| Clinical data category | ||||||||
| Clinical presentation | 8 | 3 | 3 | 2 | 2 | 0 | 1 | 3 |
| Exposure | 5 | 3 | 2 | 0 | 1 | 0 | 1 | 1 |
| Physical examination | 13 | 4 | 3 | 6 | 2 | 0 | 1 | 7 |
| Sign and symptom | 6 | 2 | 1 | 3 | 0 | 0 | 1 | 3 |
| Laboratory | 21 | 7 | 6 | 8 | 3 | 0 | 3 | 11 |
| Imaging CT | 10 | 4 | 3 | 3 | 2 | 0 | 1 | 4 |
| Treatment plan | 8 | 3 | 2 | 3 | 1 | 0 | 1 | 4 |
| Discharge outcome | 14 | 4 | 5 | 5 | 3 | 0 | 2 | 7 |
| Total | 137 | 43 | 45 | 49 | 24 | 0 | 21 | 69 |
CT=Computed tomography
After the second round of Delphi [Table 1], 45 data fields for clinical and 23 fields for the administrative category were excluded from primary MDS [Table 1]. Overall, the ultimate data fields for administrative and clinical categories were 29 and 40, respectively. In the next stage, for each finalized data field, their corresponding content was extracted from real patient medical records. After defining the information content for the fields, they were coded using selected classification or nomenclature systems (preferred codes). Then, all scattered codes were mapped to integrated codes at SNOMED-CT through MindMaple software. Tables 2 and 3 report the data classes, fields, corresponding content, data format, content definition, as well as preferred and reference codes for clinical and administrative data categories.
Table 2.
Administrative minimum data set description for information exchange of coronavirus disease-19
| Required data elements | Real case definition | |||||
|---|---|---|---|---|---|---|
| Data classes/items | Content definition | Response format | Case sample | Vocab code | Preferred codes | Reference codes |
| A. Demographical data | ||||||
| Name, surname | First/middle/last name | String | Patient name | XaLva | 371484003 | |
| Father name | First name | String | Person name | XaLva | 734006007 | |
| Age (years) | Infant: x <1 year*, child: 1 year < x <5 years*, teenage: 5 years< x <17 years*, young: 17 years< x <34 years*, middle age: 34 years < x <65 years*, aged: x >65 years* | Integer | Middle age: 58 years | RCC | X24Ai | 28288005 |
| Sex | Male*, female* | Force choice | F | RCC | X768C | 703118005 |
| National ID | Numbers range from two to ten digits with two separator dash | Integer | National ID: xx to xxx- xxxxxx-x | RCC | XE2Hj | 422549004 |
| Date of birth | yyyy/mm/dd | Date | 1962/10/17 | RCC | 9155 | 184099003 |
| Place of birth | Geographical location: Province, city, village | Forced choice and string | Iran/Tehran | RCC | XaG3t | 315446000 |
| Marital status | Single*, married*, widow*, other* | Force choice | Married | RCC | XE0oa | 87915002 |
| Employment status | Unemployed*, employed*, retired*, student*, other* | Force choice | Employed | RCC | Ua0TB | 224363007 |
| Occupation | Free text | String | EMS nurse | RCC | XaBrW | 106292003 |
| Educational level | Illiterate*, under diploma*, diploma*, bachelor*, master of science or above*, unspecified* | Forced choice and string | Received university education | RCC | Ua0Rt | 224300008 |
| Race/nationality | Iranian: Persian*, Kurdish*, Turkish*, other* | Forced choice and string | Iranian/Persia | RCC | Xa6g5 | 297553001 |
| Home address | Province-city-street- alley-house no | String | Tehran | RCC | 134Z | 433178008 |
| City-street-alley-house no | RCC | 9153 | 184097001 | |||
| Postal/zip code | Ten digit with dash | Integer | xxxxx-xxxxx | RCC | 9158 | 184102003 |
| Phone number | Ten digit with + 98 | Integer | xxxxx-xxxxx | RCC | 9158 | 824551000000105 |
| B. Admission data | ||||||
| Admission date | yyyy/mm/dd | Date | 2020/2/5 | RCC | Xa0cK | 399423000 |
| Reason for admission | Free text | String | Influenza-like symptoms | ICD10 | R68.8 | 315642008 |
| Medical record number | Six digit with two separator dashes | Integer | MRN: xx-xx-xx | RCC | Xn73J | 398225001 |
| Social security number | Nine digit with two separator dash | Integer | SSN: XXX-XX-XXXX | RCC | XagCD | 398093005 |
| Physician ID | Numbers range from two to eight digits | Integer | phys. id: xx to xxxxxxxx | RCC | Xabhz | 713578002 |
| Insurance ID | Eight digit number | Integer | Ins. id: xxxxxxxx | RCC | XE2Hj | 456281000000100 |
| C. Report Identification data | ||||||
| Report heading | COVID-19 reporting | String | Unstructured free text | RCC | Xa4H9 | 716931000000107 |
| Report ID | rep. id: xxx-x-xx | Integer | Six digit with two dash | RCC | Xbn9Z | 439272007 |
| Report Date | yyyy/mm/dd | Date | yyyy/mm/dd | RCC | Uc35Z | 399651003 |
| Reporter user ID | Personnel id: xxxx | Integer | Numbers range from three to eight digits | RCC | Xabhz | 713578002 |
| Recipient user ID | Personnel id: xxxx | Integer | Numbers range from three to eight digits | RCC | Xabhz | 713578002 |
| Reporting organization ID | Hospital ref. no: xxxx | Integer | Numbers range from two to eight digits | RCC | 9R6K | 185975009 |
| Recipient organization ID | Public health no. xxx | Integer | Numbers range from two to eight digits | RCC | XaC8K | 719051000000105 |
| Sample ID | Sample id no. xx-xx | Integer | Four digit with a separator dash | RCC | 4j33 | 719051000000105 |
RCC=Renal cell carcinoma, COVID=Coronavirus disease
Table 3.
Clinical minimum data set description for information exchange of coronavirus disease-19
| Required data elements | Real case definition | |||||
|---|---|---|---|---|---|---|
| Data classes/items | Content definition | Response format | Case sample | Vocabcode | Preferredcodes | Reference codes |
| D. Clinical presentation | ||||||
| Current existing condition | Hypertension Chronic respiratory diseases (specify type) | Select all that apply and string | Mild COPD | ICD10 | J44.8 | 313296004 |
| Diabetes | ||||||
| Coronary heart disease (specify type) | ||||||
| Cerebrovascular diseases (specify type) | ||||||
| Mental diseases (specify type) | ||||||
| Cancer (specify type) | ||||||
| HIV/AIDS infection | ||||||
| Renal diseases (specify type) | ||||||
| Liver disease | ||||||
| Other | ||||||
| Pregnancy status (if patient is a woman) | Force choice | Not pregnant | RCC | X76Qu | 60001007 | |
| Days from exposure to symptom onset | <2 days*, 2-4 days*, 4-7 days*, 1-2 weeks*, 2-4 weeks*, 1-3 months*, 3-6 months*, 6-12 months*, 1 year*< | Integer | 10 days | RCC | XaB8B | 307474000 |
| Days from illness onset to treatment | Integer | 2 days | RCC | XaB8B | 307474000 | |
| E. Exposure to casual factors | ||||||
| Exposure history | Contact/bitten with sick domestic or wild animal | Select all that apply and string | Contact with suspicious person outside wards | ICD10 CM | Z03.818 | 506901000000103 |
| Contact with suspicious person outside wards | ||||||
| Contact with patients in isolation wards | ||||||
| Contact with specimens | ||||||
| Exposure to contaminated surfaces Other | ||||||
| F. Physical examination | ||||||
| Body mass index (kg/m2) | <18.5*, between 18.5 and 24.9*, between 25 and 29.9*, >30*, unknown* | Force choice and integer | Body mass index 25-29, overweight | ICD10 | E66.9 | 162863004 |
| Respiratory rate | ≤24 breaths per min* >24 breaths per min* | Force choice and integer | 18 breath per minute | ICD10 | R06.89 | 289100008 |
| Temperature (°C) | <37.3*, 37.3-38*, 38.1-39*, >39.0* | Force choice and integer | Body temperature above 39 | ICD10 | R50.9 | 50177009 |
| Heart rate (bit/min) | <60*, between 60 and 100*, >100*, unknown* | Force choice and integer | Normal heart rate | RCC | Xa7s1 | 76863003 |
| Blood group | RH positive: A, B, AB, O RH negative: A, B, AB, O | Force choice and string | Blood group B Rh (D) positive | RCC | Xa0dT | 278150003 |
| Blood pressure (mmHg) | <120*, between 120 and 129*, between 130 and 139*, >140*, unknown* | Force choice and integer | Normal BP, 120-129 | RCC | Ua1fM | 2004005 |
| Lung examination | Clear or normal*, rales*, decreased breath sounds or dullness*, rhonchi*, wheezing*, other* | Select all that apply and string | Rhonchi present | ICD10 | R09.8 | 268929007 |
| G. Signs and symptoms | ||||||
| Asymptomatic | Yes*, no* | Force choice | Symptomatic disease | RCC | XC0v5 | 264931009 |
| If asymptomatic response is “NO,” the symptom is: | Fever Cough Dyspnea | Select all that apply and string | Dry cough Dyspnea Fever | ICD10 | R06.2 R06.8 R50.9 | 49727002 230145002 722892007 8579004 |
| weakness | Weakness | R11 | ||||
| Myalgia | ||||||
| Chest tightness or pain | ||||||
| Expectoration | ||||||
| Headache | ||||||
| Sore throat | ||||||
| Diarrhea | ||||||
| Anorexia | ||||||
| Nausea | ||||||
| Abdominal pain | ||||||
| Hemoptysis | ||||||
| Other | ||||||
| Symptom onset date | yyyy/mm/dd | Date | 2020/1/28 | RCC | XaR6r | 520191000000103 |
| H. Laboratory findings | ||||||
| Sample type | Nasopharyngeal swab | Select all that apply and | Nasopharyngeal swab | RCC | 412B | 168141000 |
| Oropharyngeal swab | string | |||||
| Broncho alveolar lavage | ||||||
| Nasopharyngeal aspirate | ||||||
| Sputum | ||||||
| Tissue (lung) biopsy | ||||||
| Serum | ||||||
| Whole blood test | ||||||
| Stool | ||||||
| Urine | ||||||
| Other | ||||||
| CBC | White blood cell count | Integer | CBC routine test | LOINC | 24317-0 | 26604007 |
| Lymphocyte count | ||||||
| Platelet count, hemoglobin Neutrophil count | ||||||
| Coagulation profiles | Prothrombin time APTT | Integer | Coagulation/bleeding tests normal | RCC | 42Q1 | 165562007 |
| D-dimer | ||||||
| Blood lipids and electrolytes | Triglyceride Total cholesterol | Integer | Serum triglycerides borderline high | RCC | 44Q3 44I2 | 442193004 166685005 |
| Low-density lipoprotein | Electrolytes normal | |||||
| Serum potassium | ||||||
| Serum sodium | ||||||
| Blood gases analysis | PaO2 | Integer | Normal blood gases | RCC | X7702 | 250544002 |
| PaO2/FiO2 | ||||||
| Lactic acid | ||||||
| PaCO | ||||||
| Liver and renal function | Creatinine Aspartate aminotransferase | Integer | Serum creatinine raised | ICD10 | R79.8 | 166717003 |
| Albumin | ||||||
| Alanine aminotransferase | ||||||
| Specialty LAB | Elisa test*, real-time PCR*, virus culture*, Other* | Select all that apply and string | Analysis using real time PCR | LOINC | 76581-8 | 444076003 |
| Sampling time | yyyy/mm/dd | Date | 2020/2/3 | RCC | 4I32 | 168149003 |
| Test time | yyyy/mm/dd | Date | 2020/2/4 | RCC | X77Vk | 252127002 |
| Sampling location | Nasal*, pharyngeal*, mouth*, lung*, blood vessel*, other* | Select all that apply and string | Nasopharyngeal | RCC | Xa0GE | 71836000 |
| Test result | Positive CoV*, negative CoV* | Force choice | Positive COVID-19 | ICD10 | R84.5 | 13320001000004109 |
| I. Imaging CT | ||||||
| Chest CT-scan | Unilateral*, bilateral* | Force choice | Bilateral chest CT-scan | ICD9 CM | 87.41 | 426827002 |
| CT features | GGO Consolidation interlobular septal thickening | Select all that apply and string | Lung consolidation | ICD10 | J18.1 | 95436008 |
| Crazy paving pattern | ||||||
| Air bronchogram | ||||||
| Spider web sign | ||||||
| Subpleuoral line | ||||||
| Bronchial wall thickening | ||||||
| Lymph node enlargement | ||||||
| Pericardial effusion | ||||||
| Plural effusion Other | ||||||
| Lung segment involvement | Average lung Dorsal of right lower | Select all that apply and | Right lower zone pneumonia | ICD10 | J18.1 | 301001009 |
| Lateral basal of right lower | string | |||||
| Posterior basal of right lower | ||||||
| Dorsal of left lower | ||||||
| Posterior basal of left lower | ||||||
| Other | ||||||
| Distribution | Sub pleural diffuse | Force choice | Pleural effusion | ICD10 | J11.1 | 81075000 |
| Per bronchial | ||||||
| Peri bronchovascular | ||||||
| Mixed | ||||||
| J. Treatment plan | ||||||
| Oxygen therapy | Noninvasive mechanical ventilator*, Invasive mechanical ventilator*, ECMO*, other* | Select all that apply and string | Noninvasive ventilation therapy | ICD9 CM | 93.90 | 784821000000105 |
| Drug therapy | Antibiotic treatment*, antifungal treatment*, antiviral treatment*, glucocorticoids*, intravenous immunoglobulin therapy*, other* | Select all that apply and string | Corticosteroid | RX- NORM | C0010137 | 79440004 |
| Complementary therapy | Yes*, no*, if yes, mention the procedure type* | Select all that apply and string | Respiratory rehabilitation | ICD9 CM | 93.99 | 790841000000106 |
| Consultation program | Mental*, occupational*, family*, social*, other* | Force choice | Mental counseling | ICD9 CM | 89.08 | 313080005 |
| K. Discharge outcome | ||||||
| Discharge date | yyyy/mm/dd | Date | 2020/2/9 | RCC | XaZuU | 442864001 |
| Discharge status | Death*, full recovery*, partial recovery*, other* | Force choice | Postdischarge follow-up | RCC | Xaat1 | 406151001 |
| If death, underlying cause of death | Related to current disease*, unrelated to current disease*, not applicable*, unknown* | Force choice | Not applicable | RCC | X90ca | 385432009 |
| If death, date of death | yyyy/mm/dd* | Date | Not applicable | RCC | X90ca | 385432009 |
| Discharge location | Home*, hospital*, other care facilities*: 1- quarantine centers, 2- nursing facility, 3- hospice care, 4-rehabilitation facility | Forced choice | Discharge to home | RCC | XaApt | 306689006 |
| Discharge Prescribed drugs | Drug name | String | Naproxen 200 mgtetracycline 250 mg | RX-NORM | C0027396 C0974349 | 416821000 324012004 |
| Date of follow up | yyyy/mm/dd | Date | 2020/2/14 | RCC | 8H8Z | 183616001 |
COPD=Chronic obstructive pulmonary disease, RCC=Renal cell carcinoma, BP=Blood pressure, CBC=Complete blood count, APTT=Activated partial thromboplastin time, PCR=Polymerase chain reaction, COVID=Coronavirus disease, CoV=Coronavirus, CT=Computed tomography, GGO=Ground-glass opacity, ECMO=Extracorporeal membrane oxygenation, LAB=Laboratory
XML schemas
XML schemas of COVID-19 provide a tools of defining the structure, content and semantics of exchange reports. The report template is divided into administrative and clinical sections. In Figure 2 presents XML based CDA framework related to COVID-19 reporting [Figure 2].
Figure 2.
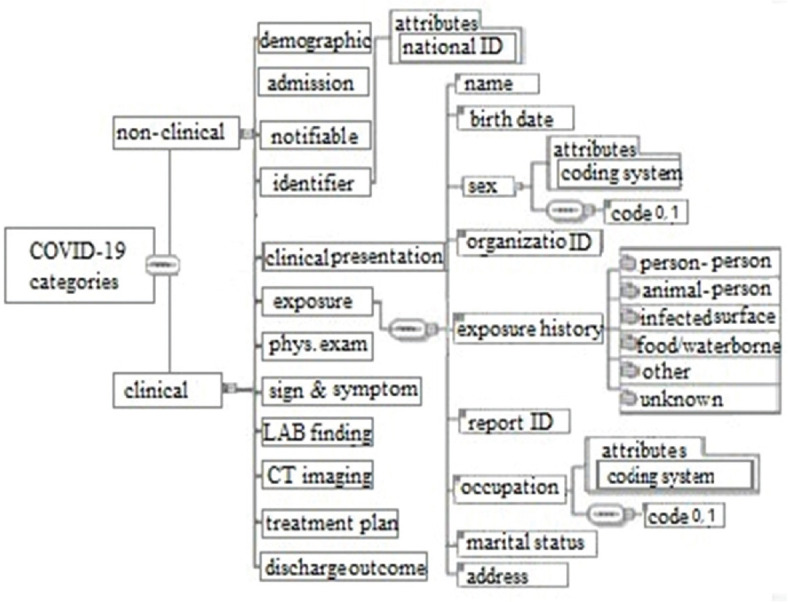
Extensive Markup Language-based Clinical Document Architecture hierarchical framework related to COVID-19 disease reporting
The HL7-CDA standard was used for standardization of the message syntax. In the CDA structure, the data field related to identification of entities was pasted into the document heading, while the CDA body contained detailed information about clinical findings [Figure 3].
Figure 3.
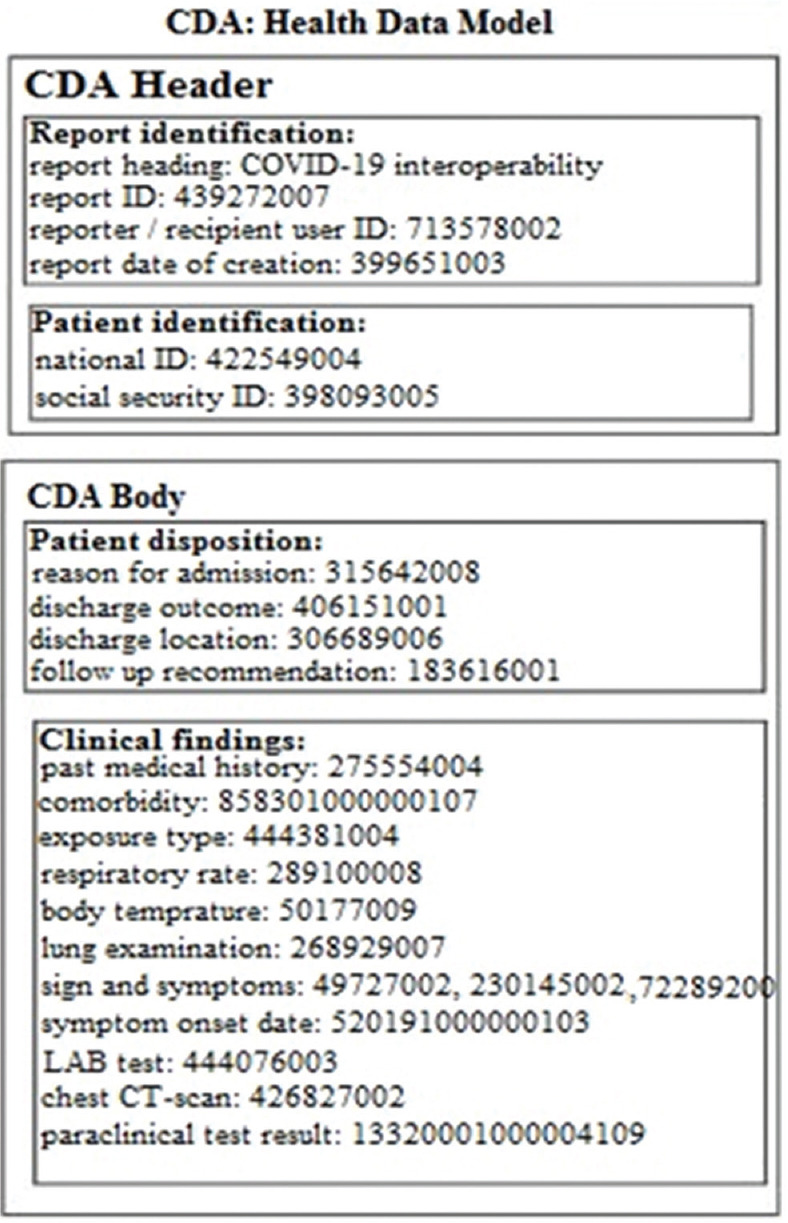
Free-text Health Level Seven-Clinical Document Architecture framework for information exchange of COVID-19 reporting
Discussion
With the widespread outbreak of COVID-19, Iran Ministry of Health and Medical Education has focused on the coordination of care and highlights the need to standardized data collection to streamline and improve the surveillance capabilities of Iranian Health system in response to this pandemic. In this regard, developing a unified and interoperable reporting framework is most effective to prompt detection and tracking of cases, investigate causes, and control a disease outbreak.[20,21,22] The purpose of MDS is to standardize the collection and reporting of a minimal amount of data as a basis for implementing any electronic systems for clinical, research, surveillance, and management purposes.[23,24,25,26] The developed MDS in this study primarily focused on PHS, whoever can be used for other applications. In this regard, we initially defined an MDS required for unified data reporting of COVID-19. Then, the structure and semantics of COVID-19 disease reporting were standardized according to HL7-CDA for the purpose of information exchange.
The quality of surveillance systems can be limited due to poor uptake or unreliable data entry process. Manual data entry is time-consuming and suffers from the inconsistent and poor-quality data structured forms. Furthermore, reports are inadequate and data are input into incorrect or erroneous fields. Thus, a reliable and friendly data entry process is crucial for capturing high quality data. Each data field should also be comprehensive so that it can be recorded in a few clicks. From a health-care provider's perspective, it is easier to analyze the data fields that are compulsory options rather than free-text data.[27,28] To compliance with data quality criteria such as data consistency and comparability in COVSS, not only a COVID-19 MDS but also more detailed categories (levels) and data formats for data capturing were defined.
New improvements in data collection instruments support the findability, accessibility, interoperability, and reusability (FAIR) of data, emphasizing the need for uniform data that can be integrated from distributed databases.[29,30,31] In this regard, this study therefore provides exchange, aggregate, and proper data management to reach FAIR data regarding COVID-19.[32]
Given the prevalence of COVID-19 in Iran,[33,34,35] the current study determined the national COVSS MDS, to collect, analyze, and report COVID-19 indicators. Each data element was mapped to common coding standards and terminologies to facilitate interoperability between various health systems at local, national, and global levels.
The COVSS MDS can be used in other countries as a main prerequisite to the implementation of the COVID-19 surveillance system. This study also highlights the benefits of standardization of COVID-19 data exchange processes which can be useful to other public health domains. Interoperable reporting for COVID-19 provides timely and reliable clinical data for measuring disease trends, efficiently applying control and prevention actions, detecting high-risk inhabitants or geographic zones, and keeping the clinical community informed through warnings, recommendations, notifies, and guidelines.[36,37,38]
Our study method had three major strengths. First of all, the proposed COVSS MDS was gathered through an extensive literature review combined with a two-round Delphi survey that benefits from evidence based and expert's wisdom in determining data elements. Second, the adoption of standard nomenclature such as SNOMED-CT is suggested for the Electronic Health Record (EHR) as it captures clinical information at the level of details required by clinicians for care provision in most health-care disciplines and settings. Finally, we leveraged HL7-CDA, as a standard for the exchange of clinical documents, which should be readable by computers and humans. HL7 CDA is an XML-based standard which has a simple and very flexible text format for structuring and exchanging information on the Web environment.[39,40]
Given some of the unfamiliar aspects of this novel outbreak, we recommend the development of conceptual models of surveillance systems and conducting a pilot study including a further Delphi stage prior to refine some data categories. In addition, this MDS may need to be appraised from the perspectives of a greater group of clinical and public health professionals to be applicable in a nationwide. Further, this study provides COVID-19 interoperable reporting framework from a data management perspective, but its technological aspects need to be resolved which are beyond our discussions in this article.
Conclusion
An effective COVID-19 surveillance system requires complete and timely information to guide fully informed decisions to reduce the further spread of disease by taking early preventive measures. The template presented in this study can enable interoperability across many clinical and public health information systems that populate the COVID-19 surveillance system. The main output of the proposed template supports collaborations among various healthcare providers and public health agencies in patient care management as well as research or public health purposes. Given some of the unfamiliar aspects of this novel outbreak, we recommend the development of conceptual models of surveillance systems and conducting a pilot study including a further Delphi stage prior to refine some data categories.
Financial support and sponsorship
This research project has been financially supported by Abadan Faculty of Medical Sciences (Iran) under contract number of 98U749.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This article is the result of a research project approved by the research committee at Abadan Faculty of Medical Sciences (Iran) (Ethic code number: IR. ABADANUMS. REC.1398.109). The authors thank all of the clinical and health information management experts that cooperated with them to complete questionnaire.
References
- 1.Jung S-m, Akhmetzhanov AR, Hayashi K, Linton NM, Yang Y, Yuan B, et al. Real-time estimation of the risk of death from novel coronavirus (covid-19) infection: Inference using exported cases. J Clin Med. 2020;9:523. doi: 10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–4. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C, Chen X, Cai Y, Zhou X, Xu S, Huang H, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Int Med. 2020 Mar 13; doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;3(55) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, et al. Presumed asymptomatic carrier transmission of COVID-19 2020 Apr 14. 323(14):1406–7. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung SM, et al. Epidemiological characteristics of novel coronavirus infection: A statistical analysis of publicly available case data medRxiv. 2020 doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Organization WH. Coronavirus Disease 2019 (COVID-19): Situation Report. 2020:45. [Google Scholar]
- 8.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards CL, Iademarco MF, Atkinson D, Pinner RW, Yoon P, Mac Kenzie WR, et al. Advances in public health surveillance and information dissemination at the centers for disease control and prevention. Public Health Rep. 2017;132:403–10. doi: 10.1177/0033354917709542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon BE, Rahurkar S, Ho Y, Arno JN. Reliability of administrative data to identify sexually transmitted infections for population health: A systematic review. BMJ Health Care Inform. 2019 Aug 1;26(1) doi: 10.1136/bmjhci-2019-100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Streefkerk HR, Verkooijen RP, Bramer WM, Verbrugh HA. Electronically assisted surveillance systems of healthcare-associated infections: A systematic review. Euro Surveill. 2020 Jan 16;25(2) doi: 10.2807/1560-7917.ES.2020.25.2.1900321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allam Z, Jones DS. On the coronavirus (COVID-19) outbreak and the smart City Network: Universal Data Sharing Standards Coupled with Artificial Intelligence (AI) to Benefit Urban Health Monitoring and Management Healthcare (Basel) 2020 Feb 22;8(1) doi: 10.3390/healthcare8010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safdari R, Ghazi Saeedi M, Masoumi-Asl H, Rezaei-Hachesu P, Mirnia K, Mohammadzadeh N, et al. National minimum data set for antimicrobial resistance management: Toward global surveillance system. Iran J Med Sci. 2018;43:494–505. [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia MC, Garrett NY, Singletary V, Brown S, Hennessy-Burt T, Haney G, et al. An assessment of information exchange practices, challenges and opportunities to support US disease surveillance in three states. J Public Health Manag Practice. 2018;24:546. doi: 10.1097/PHH.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gansel X, Mary M, van Belkum A. Semantic data interoperability, digital medicine, and e-health in infectious disease management: A review. Eur J Clin Microbiol Infect Dis. 2019;38:1023–34. doi: 10.1007/s10096-019-03501-6. [DOI] [PubMed] [Google Scholar]
- 16.Pilot E, Roa R, Jena B, Kauhl B, Krafft T, Murthy G. Towards sustainable public health surveillance in India: Using routinely collected electronic emergency medical service data for early warning of infectious diseases. Sustainability. 2017;9:604. [Google Scholar]
- 17.Gazzarata R, Monteverde ME, Ruggiero C, Maggi N, Palmieri D, Parruti G, et al. Healthcare associated infections: An interoperable infrastructure for multidrug resistant organism surveillance. Int J Environ Res Public Health. 2020 Jan;17(2):465. doi: 10.3390/ijerph17020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheikhali SA, Abdallat M, Mabdalla S, Al Qaseer B, Khorma R, Malik M, et al. Design and implementation of a national public health surveillance system in Jordan. Int J Med Inform. 2016;88:58–61. doi: 10.1016/j.ijmedinf.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cato KD, Cohen B, Larson E. Data elements and validation methods used for electronic surveillance of health care-associated infections: A systematic review. Am J Infect Control. 2015;43:600–5. doi: 10.1016/j.ajic.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raeisi A, Tabrizi JS, Gouya MM. IR of Iran national mobilization against COVID-19 Epidemic. Arch Iran Med. 2020;23:216–9. doi: 10.34172/aim.2020.01. [DOI] [PubMed] [Google Scholar]
- 21.Mounesan L, Eybpoosh S, Haghdoost A, Moradi G, Mostafavi E. Is reporting many cases of COVID-19 in Iran due to strength or weakness of Iran's health system? Iran J Microbiol. 2020;12:73–6. [PMC free article] [PubMed] [Google Scholar]
- 22.Moradzadeh R. The challenges and considerations of community-based preparedness at the onset of COVID-19 outbreak in Iran, 2020. Epidemiol Infect. 2020;148:e82. doi: 10.1017/S0950268820000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanbehzadeh M, Ahmadi M. Identification of the necessary data elements to report AIDS: A systematic review. Electron Physician. 2017;9:5920–31. doi: 10.19082/5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazemi-Arpanahi H, Vasheghani-Farahani A, Baradaran A, Mohammadzadeh N, Ghazisaeedi M. Developing a minimum data set (MDS) for cardiac electronic implantable devices implantation. Acta Inform Med. 2018;26:164–8. doi: 10.5455/aim.2018.26.164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazemi-Arpanahi H, Vasheghani-Farahani A, Baradaran A, Ghazisaeedi M, Mohammadzadeh N, Bostan H. Development of a minimum data set for cardiac electrophysiology study ablation. J Educ Health Promot. 2019;8:101. doi: 10.4103/jehp.jehp_232_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baunsgaard CB, Chhabra H, Harvey L, Savic G, Sisto SA, Qureshi F, et al. Reliability of the international spinal cord injury musculoskeletal basic data set. Spinal cord. 2016;54:1105–13. doi: 10.1038/sc.2016.42. [DOI] [PubMed] [Google Scholar]
- 27.Davey CJ, Slade SV, Shickle D. A proposed minimum data set for international primary care optometry: A modified Delphi study. Ophthalmic Physiol Opt. 2017;37:428–39. doi: 10.1111/opo.12372. [DOI] [PubMed] [Google Scholar]
- 28.Revere D, Hills RH, Dixon BE, Gibson PJ, Grannis SJ. Notifiable condition reporting practices: Implications for public health agency participation in a health information exchange. BMC Public Health. 2017;17:247. doi: 10.1186/s12889-017-4156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016:3. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haywood KL, Griffin XL, Achten J, Costa ML. Developing a core outcome set for hip fracture trials. Bone Joint J. 2014;96-B:1016–23. doi: 10.1302/0301-620X.96B8.33766. [DOI] [PubMed] [Google Scholar]
- 31.Lutomski JE, Baars MA, Schalk BW, Boter H, Buurman BM, den Elzen WP, et al. The development of the older persons and informal caregivers survey minimum dataset (TOPICS-MDS): A large-scale data sharing initiative. PloS One. 2013;8:e81673. doi: 10.1371/journal.pone.0081673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley WT, Glasgow RE, Etheredge L, Abernethy AP. Rapid, responsive, relevant (R3) research: A call for a rapid learning health research enterprise. Clin Translat Med. 2013;2:10. doi: 10.1186/2001-1326-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reza G, Fatemeh H. Covid-19 and Iran: Swimming with hands tied! Swiss Med Weekly. 2020 Apr 7;150(1516) doi: 10.4414/smw.2020.20242. [DOI] [PubMed] [Google Scholar]
- 34.Zandifar A, Badrfam R. Fighting COVID-19 in Iran; Economic challenges ahead. Arch Iran Med. 2020;23:284. doi: 10.34172/aim.2020.14. [DOI] [PubMed] [Google Scholar]
- 35.Raoofi A, Takian A, Akbari Sari A, Olyaeemanesh A, Haghighi H, Aarabi M. COVID-19 Pandemic and Comparative Health Policy Learning in Iran. Arch Iran Med. 2020;23:220–34. doi: 10.34172/aim.2020.02. [DOI] [PubMed] [Google Scholar]
- 36.Gong M, Liu L, Sun X, Yang Y, Wang S, Zhu H. Cloud-based system for effective surveillance and control of COVID-19: Useful experiences from Hubei, China. J Med Internet Res. 2020;22:e18948. doi: 10.2196/18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desjardins MR, Hohl A, Delmelle EM. Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: Detecting and evaluating emerging clusters. Appl Geography. 2020:118. doi: 10.1016/j.apgeog.2020.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foddai A, Lindberg A, Lubroth J, Ellis-Iversen J. Surveillance to improve evidence for community control decisions during the COVID-19 pandemic– Opening the animal epidemic toolbox for Public Health. One Health. 2020:9. doi: 10.1016/j.onehlt.2020.100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu D, Wang X, Pan F, Xu Y, Yang P, Rao K. Web-based infectious disease reporting using XML forms. Int J Med Inform. 2008;77:630–40. doi: 10.1016/j.ijmedinf.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Kokkinakis I, Selby K, Favrat B, Genton B, Cornuz J. Covid-19 diagnosis: Clinical recommendations and performance of nasopharyngeal swab-PCR. Rev Med Suisse. 2020;16:699–701. [PubMed] [Google Scholar]


