Abstract
The SARS-CoV-2 pandemic has caused extreme human suffering and economic harm. We generated and characterized a new mouse-adapted SARS-CoV-2 virus that captures multiple aspects of severe COVID-19 disease in standard laboratory mice. This SARS-CoV-2 model exhibits the spectrum of morbidity and mortality of COVID-19 disease as well as aspects of host genetics, age, cellular tropisms, elevated Th1 cytokines, and loss of surfactant expression and pulmonary function linked to pathological features of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). This model can rapidly access existing mouse resources to elucidate the role of host genetics, underlying molecular mechanisms governing SARS-CoV-2 pathogenesis, and the protective or pathogenic immune responses related to disease severity. The model promises to provide a robust platform for studies of ALI and ARDS to evaluate vaccine and antiviral drug performance, including in the most vulnerable populations (i.e., the aged) using standard laboratory mice.
Keywords: SARS-CoV-2, COVID-19, animal models, mouse-adaptation, acute lung injury, acute respiratory distress syndrome, interferon, vaccines
Graphical Abstract
Leist et al. present a mouse model for COVID-19 by serially passaging human SARS-CoV-2 in vivo to create an evolution-selected lethal mouse-adapted virus variant, called MA10. MA10 shows a dose- and age-related increase in pathogenesis in standard laboratory mice and recapitulates key features of COVID-19 in humans.
Introduction
Zoonotic coronaviruses (CoVs) are responsible for three epidemics in the 21st century, including severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and the ongoing Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012. In December 2019, a third novel CoV designated SARS-CoV-2 emerged (Zhou et al., 2020b; Zhu et al., 2020) and has resulted in a worldwide pandemic with over 25 million cases and over 850,000 deaths in over 220 countries (Dong et al., 2020). SARS-CoV-2 infection results in a complex clinical syndrome, designated Coronavirus Disease 2019 (COVID-19), that causes a range of clinical symptoms from mild to a severe disease associated with acute lung injury (ALI) and the acute respiratory distress syndrome (ARDS) (Guan et al., 2020). SARS-CoV-2 infection can also produce strokes, cardiac pathology, gastrointestinal disease, coagulopathy, and a hyperinflammatory shock syndrome (Mao et al., 2020; Wichmann et al., 2020; Cheung et al., 2020). The elderly, and those with underlying co-morbidities, are at increased risk of severe COVID-19 (Zhou et al., 2020a) and death is most commonly linked to respiratory failure due to ARDS (Rothan and Byrareddy, 2020; Li and Ma, 2020). In fact, the mortality rate from COVID-19 ARDS approaches 40%–50%, perhaps associated with a “cytokine storm” characterized by elevated levels of interferon (IFN)-γ, interleukin (IL)-18, transforming growth factor β (TGF-β), IL-6, IP-10, monocyte chemoattractant protein-1 (MCP-1), monokine-induced by interferon-γ (MIG), and IL-8 (Sinha et al., 2020).
SARS-CoV-2 uses the human angiotensin-converting enzyme 2 (hACE2) as a receptor for docking and entry into cells but is incapable of using the murine ortholog (mACE2) receptor (Letko et al., 2020; Zhou et al., 2020b). To generate SARS-CoV-2 mouse models, several groups have developed transgenic mouse lines expressing hACE2, utilizing a variety of exogenous (Jiang et al., 2020; Bao et al., 2020) or murine promoters (Sun et al., 2020b), or by transduction using adenovirus (Sun et al., 2020a; Hassan et al., 2020) or adeno-associated virus vectors (Israelow et al., 2020), to generate productive infections. Although each system has certain advantages (e.g., speed of development), infection in vector-mediated or transgenic overexpression models typically causes a mild alveolitis in the lung and/or progression to fatal encephalitis (Menachery et al., 2016; McCray et al., 2007; Butler et al., 2006; Jacomy et al., 2006), rarely recapitulating the severe lung disease that is one of the hallmarks of COVID-19 in humans, and/or produce a fatal encephalitis. As such, it is critical to develop models that use standard laboratory mice, reproduce age-dependent SARS-CoV-2 susceptibility, target nasal epithelia and alveolar pneumocytes, and develop the relevant pulmonary lesions of acute lung injury (including pneumonitis, edema, necrotic debris, and hyaline membrane formation) that are consistent with progression to ARDS. Such models, especially if available in standard laboratory mice, will accelerate studies of COVID-19 immune pathologies, the function of host genes in regulating disease progression, and will provide a high-throughput screening platform for evaluating antiviral drugs and vaccines.
In this study, we describe a lethal mouse model of mouse-adapted SARS-CoV-2 (defined as SARS-CoV-2 MA10) pathogenesis that recapitulates the age-related disease severity observed in humans, ALI/ARDS, and death in wild-type BALB/c mice. Like human infections, SARS-CoV-2 MA10 infection targets the conducting and distal airways, including airway epithelial cells and AT2 cells in the terminal bronchi and alveoli, and replicates in nasal olfactory epithelium sustentacular cells and Bowman’s glands. The SARS-CoV-2 MA10 infection in BALB/c mice induces lung disease characterized by damaged airway epithelium, exfoliated cells in small airways, fibrin deposition, occasional hyaline membrane formation, pulmonary edema, surfactant expression loss, and congestion that can progress to ARDS, especially in aged animals. Many Th1 proinflammatory cytokines were elevated after infection, including IL-6. Interestingly, SARS-CoV-2 MA10 infection of C57BL/6J mice caused a milder phenotype, whereas infection of immunodeficient type I and II interferon receptor double knockout C57BL/6J mice resulted in severe weight loss and morbidity. Notably, mice vaccinated with viral vector-delivered SARS-CoV-2 spike were protected from clinical disease and infection of the lung. The development of SARS-CoV-2 MA10 provides a much-needed standard laboratory mouse model that recapitulates the age-related severity spectrum and acute lung injury phenotype observed in human SARS-CoV-2 infections. It also provides a robust model to mechanistically address novel questions in COVID-19 immunity, pathogenesis, and vaccine and antiviral drug performance in wild-type and genetically modified standard laboratory mice.
Results
Adaptation of SARS-CoV-2 MA via Serial Passaging In Vivo
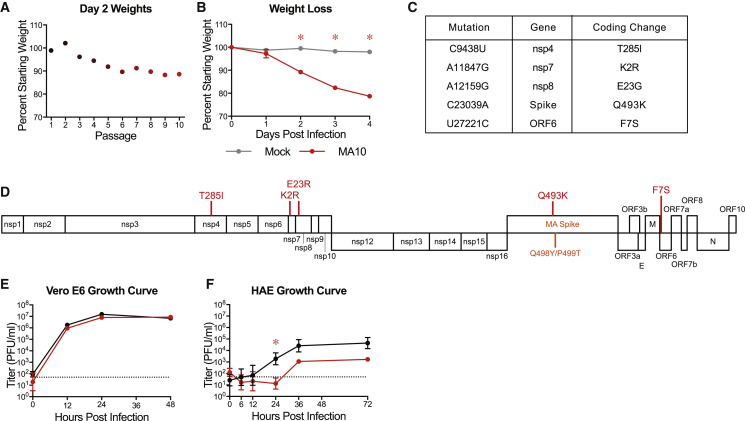
We previously developed a recombinant mouse adapted strain of SARS-CoV-2 (SARS-CoV-2 MA) capable of utilizing mACE2 for viral entry by remodeling the spike and receptor binding interface via reverse genetics (Dinnon et al., 2020). Although SARS-CoV-2 MA mediated infection of wild-type mice, young adult mice did not display the major clinical manifestations or hallmarks of ALI (Dinnon et al., 2020). To improve the model, we used experimental evolution in vivo via serial passage of SARS-CoV-2 MA in the lungs of young adult BALB/cAnNHsd mice (herein referred to as “BALB/c” mice) every 2 days to select for more virulent strains (Roberts et al., 2007). With passage, we observed a linear decrease in body weight over time achieving greater than 10% body weight loss on 2 days post infection (dpi) by passage ten (P10) (Figure 1 A). We confirmed the virulence of the virus population generated at P10 using a plaque purified clonal isolate (SARS-CoV-2 MA10) from this passage in young adult BALB/c mice (Figure 1B). Deep sequencing of mouse lung’s total RNA from the 10 passages, plaque purified SARS-CoV-2 MA10, and four additional plaque purified passage 10 viruses was performed to identify the changes responsible for the increased pathogenicity and rare variants. In addition to the spike Q498Y/P499T substitutions engineered into the parental SARS-CoV-2 MA, SARS-CoV-2 MA10 included 5 additional nucleotide changes, all resulting in nonsynonymous coding changes (Figures 1C and 1D; Table S1). These mutations emerged in an ordered fashion and include changes in nonstructural protein 4 (nsp4) (C9438T), nsp7 (A11847G), nsp8 (A12159G), spike (S; C23039A), and open reading frame 6 (ORF6; T27221C). Some sequence heterogeneity was observed across the plaque purified viruses, although SARS-CoV-2 MA10 had the fewest mutations and most represented the viral population found at passage 10 (Table S1). The SARS CoV-2 MA10 maintained the ability to utilize non-human primate ACE2 and replicated and formed plaques in Vero E6 cells (Figure 1E), consistent with utility for viral propagation and titration. Importantly, SARS-CoV-2 MA10 was also attenuated compared to wild-type SARS-CoV-2 (SARS-CoV-2 WT) in primary human bronchiolar epithelial cells (HBEs) (Figure 1F), suggesting decreased fitness in human cells. Collectively, these observations led to the choice of SARS-CoV-2 MA10 for subsequent studies.
Figure 1.
SARS-CoV-2 MA Increases in Pathogenicity following Serial In Vivo Passaging in Mice
(A) Percent starting weight at 2 dpi of mice throughout serial passage of SARS-CoV-2 MA10 in 10- to 12-week-old BALB/c mice infected with 105 PFU SARS-CoV-2 MA at passage 1, or blind titer for passages 2–10.
(B) 10-week-old BALB/c mice were mock-infected with PBS or infected with 105 PFU of plaque-purified virus from passage 10 in (A), SARS-CoV-2 MA10, and monitored for weight loss. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons.
(C) Table of mouse adaptations present in plaque purified SARS-CoV-2 MA10 relative to parental SARS-CoV-2 MA. WT, wild type; nsp, nonstructural protein; ORF, open reading frame.
(D) Schematic of SARS-CoV-2 genome with locations of mouse adaptations from (C) shown.
(E and F) Single step growth curve of SARS-CoV-2 WT and SARS-CoV-2 MA10 in Vero E6 cells (E) or differentiated primary human bronchiolar airway epithelial cells (HBE) (F). n = 3 for each group, sampled serially. Dotted line represents limit of detection. Log transformed data were analyzed by 2-factor ANOVA followed by Sidak’s multiple corrections. Error bars represent SEM about the mean for (A) and (B) and SD about the mean for (E) and (F). ∗p < 0.05.
See also Table S1.
SARS-CoV-2 MA10 Causes Acute Lung Injury in Young BALB/c Mice
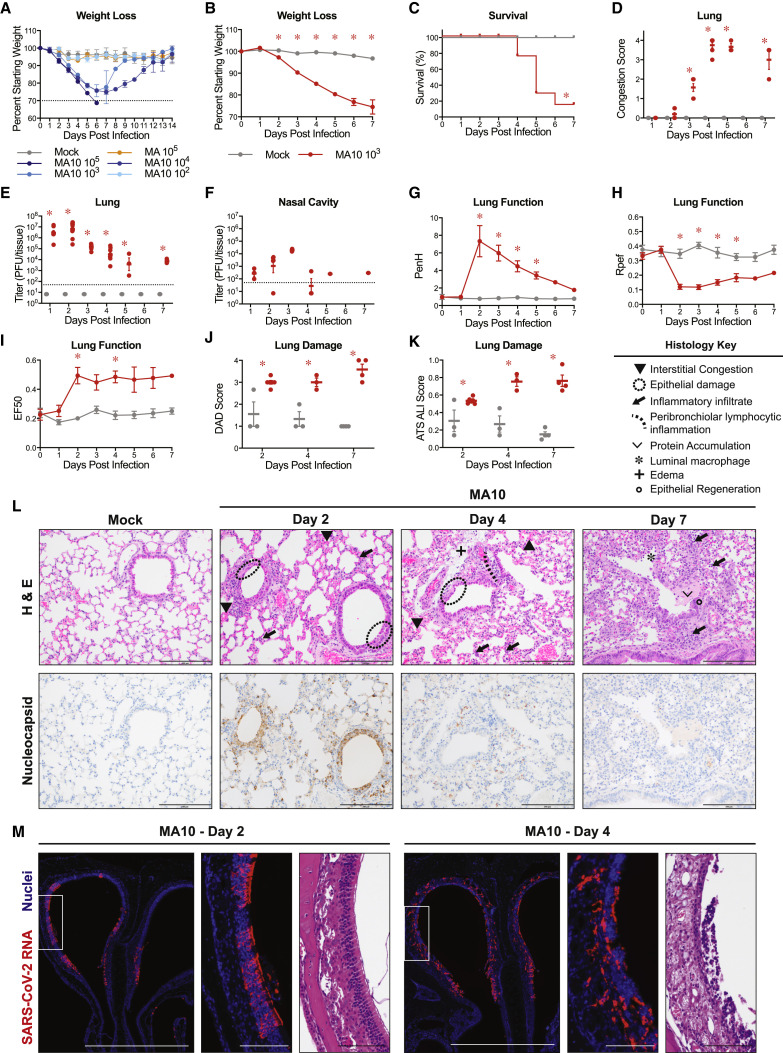
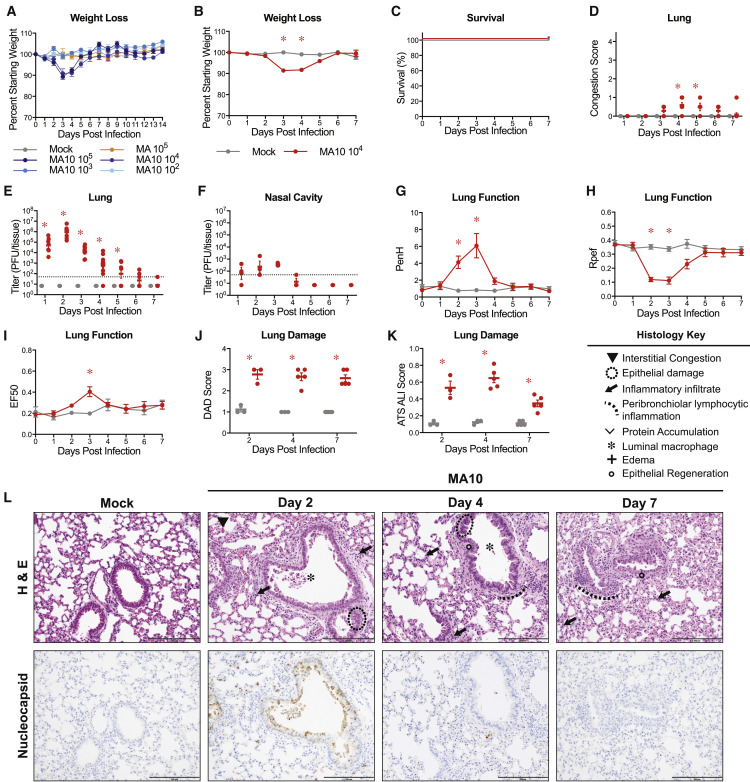
To gain insight into the dose-dependent pathogenic potential of SARS-CoV-2 MA10, we performed dose ranging studies in 10-week-old BALB/c mice infected with either PBS (mock), 105 plaque-forming unit (PFU) of the parental SARS-CoV-2 MA, or 102, 103, 104, and 105 PFU SARS-CoV-2 MA10. We observed a dose-dependent increase in morbidity and mortality over the course of 14 days with SARS-CoV-2 MA10 (Figure 2 A). Mortality rates of 20% and 60% were recorded for infection with 104 and 105 PFU, respectively. Notably, infection with 102 PFU of SARS-CoV-2 MA10 produced an increased weight loss as compared to 105 PFU infections with the parental SARS-CoV-2 MA strain, highlighting the increased pathogenicity gained through passaging. To best capture severe disease phenotypes without excessive mortality, we proceeded with 104 PFU of SARS-CoV-2 MA10 as the standard infection dose for young adult BALB/c mice.
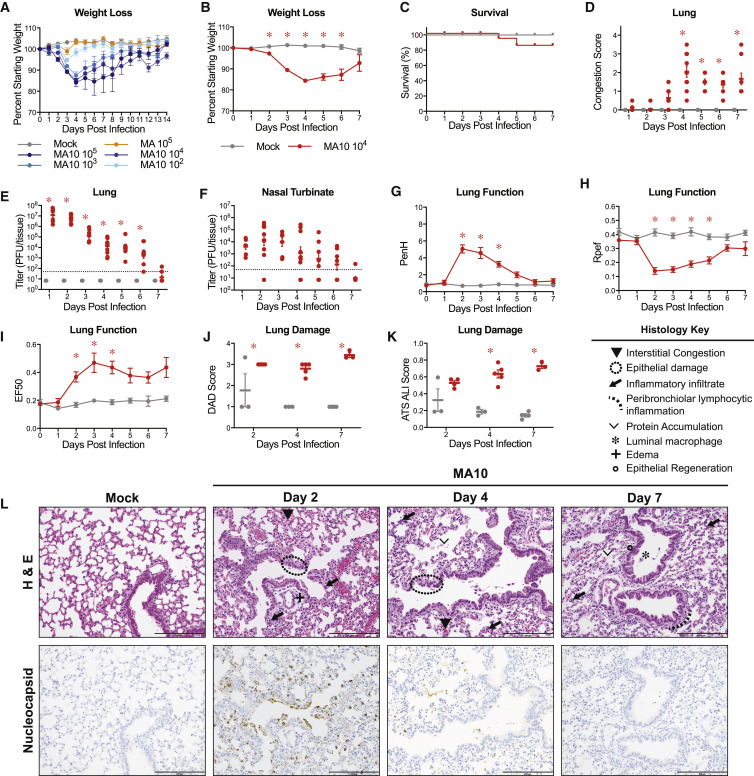
Figure 2.
SARS-CoV-2 MA10 Causes Acute Lung Injury in Young Adult BALB/c Mice
(A) 10-week-old female BALB/c mice (n = 5 per group) were mock-infected, infected with 105 PFU SARS-CoV-2 MA, or 102, 103, 104, and 105 PFU SARS-CoV-2 MA10.
(B–L) 10-week-old female BALB/c mice were mock-infected (n = 47) or infected with 104 PFU SARS-CoV-2 MA10 (n = 59).
(B) Percent starting weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons.
(C) Survival rate.
(D) Gross lung congestion score. Data analyzed by 2-factor ANOVA followed by Sidak’s multiple comparisons.
(E) Viral lung titer (mock-infected: 1 dpi: n = 6, 2 dpi: n = 7, 3 dpi: n = 6, 4 dpi: n = 7, 5 dpi: n = 6, 6 dpi: n = 6, 7 dpi: n = 9; SARS-CoV-2 MA10-infected: 1 dpi: n = 7, 2 dpi: n = 10, 3 dpi: n = 7, 4 dpi: n = 10, 5 dpi: n = 7, 6 dpi: n = 7, 7 dpi: n = 9). Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed as in (D).
(F) Viral nasal cavity titer. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection.
(G–I) Whole body plethysmography analysis of lung function parameters (10 mice per group at 0 dpi): PenH (G), Rpef (H), and EF50 (I). Data analyzed as in (B).
(J and K) Blinded histopathological evaluation of lung damage using DAD scoring (J) and ATS ALI scoring (K) systems on days 2, 4, and 7 after mock or SARS-CoV-2 MA10 infection. Data analyzed as in (D).
(L) Representative 200× images of lungs from mock and SARS-CoV-2 MA10-infected mice from (J) and (K). H&E shown in the top panels. Bottom panels show immunohistochemistry (IHC) labeling against SARS-CoV-2 nucleocapsid, counterstained with hematoxylin. Scale bars represent 200 μm. All error bars represent SEM about the mean. ∗p < 0.05.
To characterize the pathogenesis of SARS-CoV-2 MA10 in young BALB/c mice, we examined the kinetics of disease in mice through 7 days post infection using an intranasal inoculating dose of 104 PFU. SARS-CoV-2 MA10-infected mice rapidly lost weight and reached maximum weight loss at day 4 (losing 16% of starting weight) (Figure 2B). At 5 dpi, the weight loss trajectories of infected mice diverged, with many mice recovering body weight juxtaposed to mice that continued to lose weight, collectively resulting in a ∼15% mortality rate (Figure 2C). At the time of necropsy, acute stage lung damage was noted grossly as firm, red, heavy lobes that were scored based on the extent of congestion-related discoloration (Sheahan et al., 2020a) (indicative of edema and diffuse alveolar damage) that peaked at 4 dpi and remained high through 7 dpi (Figure 2D). Virus replication in the lung peaked 1–2 dpi and was absent in most surviving mice by 7 dpi (Figure 2E). Viral replication in the upper respiratory tract (measured by viral titer in the nasal cavity) remained high on 1–3 dpi but was non-detectable in most mice by 5 dpi (Figure 2F).
To gain insight into the impact of infection on lung physiology, pulmonary function was measured over time via whole body plethysmography (WBP). As compared to control mice, infected mice exhibited a loss in pulmonary function as indicated by significant changes in PenH and Rpef, measures of airway obstruction, and EF50, a measurement of exhalation flow rate (Figures 2G–2I).
Histopathologic analyses at 2, 4, and 7 dpi revealed early multifocal damage to conducting airway epithelia (including bronchioles) that corresponded to viral antigen staining, which was intense on 2 dpi, waned by 4 dpi, and was absent by 7 dpi (Figure 2J). Often, bronchial damage progressed to segmental epithelial denudation with an accumulation of inflammatory cells, sloughed epithelial cells, cellular debris, fibrin deposition, and plasma proteins in the airway lumens. Later post-SARS-CoV-2 MA10 infection, airway epithelia became hyperplastic with regeneration. The distal alveolar ducts and sacs were markedly altered by infection, displaying hallmarks of diffuse alveolar damage (DAD) and multifocal positive labeling of pneumocytes for viral antigen at early time points after infection. Histologic changes included hypercellular thickening of the alveolar septae caused by infiltrating immune cells, pneumocyte degeneration and necrosis, congestion of small vessels and capillaries, endothelial activation, increased neutrophils with extravasation, exudation of proteinaceous fluid and fibrin with occasional organization into hyaline membranes, and increased numbers of alveolar macrophages. Although later time points featured increased numbers of lymphocytes organizing around bronchioles, lymphocytic cuffing was not a prominent pathologic feature in comparison to findings induced by other respiratory viral pathogens. Importantly, the most severe, lingering damage over the time course was in the alveolar region.
The pathology of SARS-CoV-2 MA10-infected lungs was blindly quantified utilizing two metrics of ALI (Schmidt et al., 2018; Matute-Bello et al., 2011). First, diffuse alveolar damage (DAD) was assessed based on the degree of cellular sloughing and necrosis. SARS-CoV-2 MA10 induced DAD as early as 2 dpi and was maintained through 7 dpi (Figure 2J). Second, the American Thoracic Society (ATS) has generated a small animal model ALI scoring scheme that assesses neutrophil presence in the interstitium and alveolar space, hyaline membrane formation, protein accumulation, and alveolar septal thickening. Consistent with DAD scores, ATS ALI scores were increased in SARS-CoV-2 MA10-infected mice at 2 dpi and increased through 7 dpi (Figure 2K). Immunohistochemistry (IHC) staining for viral nucleocapsid revealed intense staining at 2 dpi and lack of staining by 7 dpi (Figure 2L), consistent with the lung viral titer data (Figure 2D). At 2 dpi, viral antigen was detected in conducting airway epithelia and in the alveoli, consistent with alveolar type II pneumocyte distribution patterns.
Increased Morbidity and Mortality in Old Mice after SARS-CoV-2 MA10 Infection
Because SARS-CoV-2 and other emerging human coronaviruses exhibit an age-dependent increase in disease severity, we investigated whether SARS-CoV-2 MA10 infection of aged mice resulted in an increased disease severity. In comparison to young mice, 1-year-old mice were highly susceptible to SARS-CoV-2 MA10, with high morbidity and nearly 100% mortality when infected with 104 and 105 PFU (Figure 3 A). Although mice infected with 103 PFU rapidly lost weight with very few animals surviving, those infected with 102 PFU did not exhibit disease signs and all survived, suggesting a threshold of virus >102 was necessary to cause significant disease in 1-year-old mice. Accordingly, we selected the lowest dose that caused severe disease (103 PFU) as the standard infection dose for 1-year-old mice. With this dose, the kinetics of weight loss were similar to young BALB/c mice. However, unlike infected young adult mice, all aged mice continued to lose weight over time and ultimately lost 30% of their starting weight, succumbing to infection and/or reaching the criteria for humane euthanasia (Figure 3B). Overall, we observed increased mortality starting on day 4 after infection with only ∼15% survival by day 7 (Figure 3C). Thus, data presented at late time points such as 6 or 7 dpi are biased toward rare survivors.
Figure 3.
SARS-CoV-2 MA10 Disease Is Severely Exacerbated in Old Mice
(A) 1-year old female BALB/c mice (n = 5 per group) were mock-infected (gray), infected with 105 PFU SARS-CoV-2 MA, or 102, 103, 104, and 105 PFU SARS-CoV-2 MA10. Dotted line represents 70% starting body weight.
(B–L) 1-year old female BALB/c mice were mock-infected (n = 51) or infected with 104 PFU SARS-CoV-2 MA10 (n = 65).
(B) Percent starting weight. Dotted line represents 70% starting body weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons.
(C) Survival rate. Analyzed by log-rank test.
(D) Gross lung congestion score. Data analyzed by 2-factor ANOVA followed by Sidak’s multiple comparisons.
(E) Lung viral titer (mock-infected: 1 dpi: n = 6, 2 dpi, n = 7; 3 dpi, n = 6; 4 dpi, n = 7; 5 dpi, n = 6; 6 dpi, n = 6; 7 dpi,: n = 13; SARS-CoV-2 MA10-infected: 1 dpi, n = 6; 2 dpi, n = 10; 3 dpi, n = 7; 4 dpi, n = 8; 5 dpi, n = 1; 6 dpi, n = 2; 7 dpi, n = 3.) Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed as in (D).
(F) Viral nasal cavity titer. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection.
(G–I) Whole body plethysmography analysis of lung function parameters (10 mice per group at 0 dpi): PenH (G), Rpef (H), and EF50 (I). Data analyzed as in (B).
(J and K) Blinded histopathological evaluation of lung damage using DAD scoring (J) and ATS ALI scoring (K) systems on days 2, 4, and 7 after mock or SARS-CoV-2 MA10 infection. Data analyzed as in (D).
(L) Representative 200× images of lungs from mock and SARS-CoV-2 MA10-infected mice from (J) and (K). H&E is shown in the top panels. Bottom panels show immunohistochemistry (IHC) labeling against SARS-CoV-2 nucleocapsid, counterstained with hematoxylin. Scale bars represent 200 μm.
(M) Representative in situ hybridization images of viral RNA in nasal cavity from SARS-CoV-2 MA10-infected mice. Scale bar represents 100 μm. All error bars represent SEM about the mean. ∗p < 0.05.
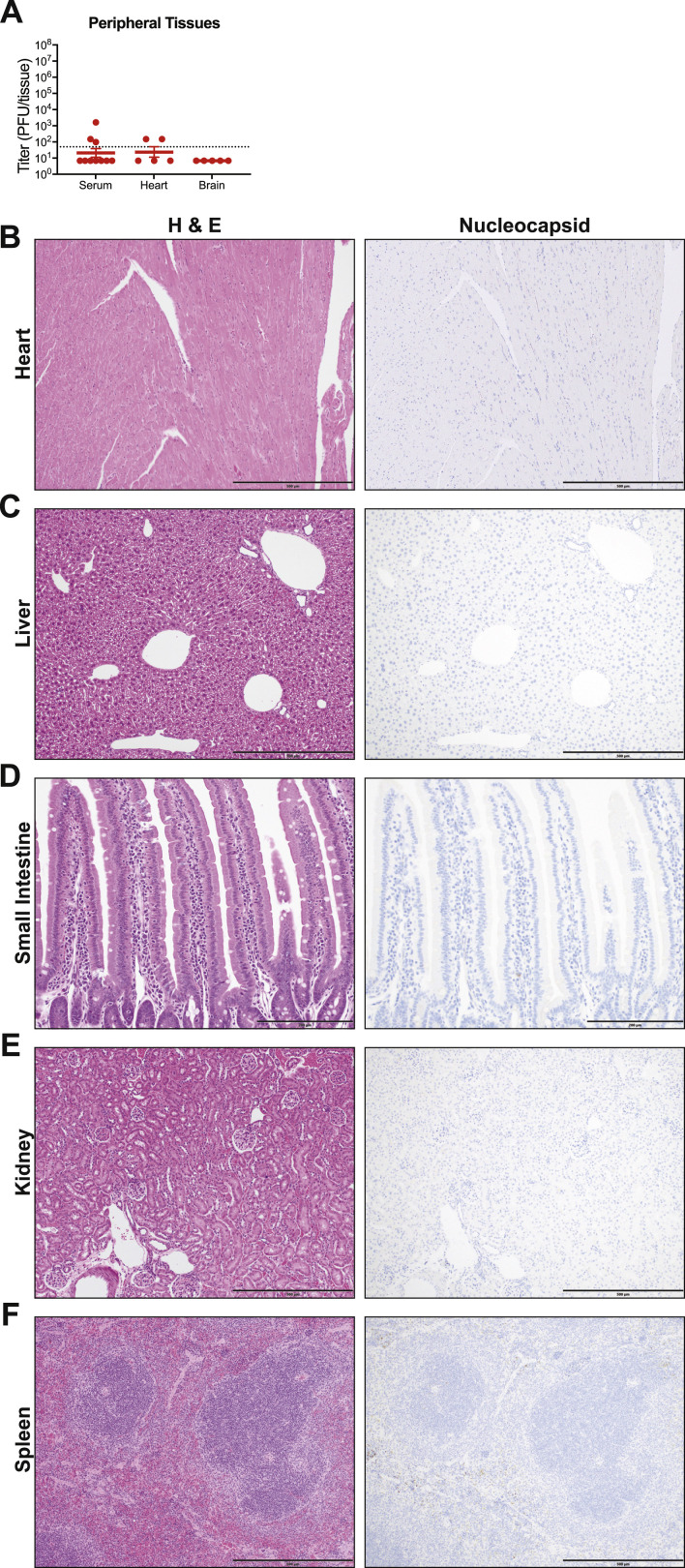
Gross pathological evaluations at necropsy revealed macroscopically detectable discoloration of lung tissue that achieved maximal severity on days 4 and 5 after infection (Figure 3D). Virus replication in aged mice peaked 1 to 2 dpi (5.3 × 106 PFU/tissue and 1.2 × 107 PFU/tissue, respectively), values similar to young adult mice. In contrast with young adult mice, in which virus was cleared by 7 dpi, significant levels of infectious virus remained in the lungs of aged mice at later time points (Figure 3E). Low levels of infectious virus were present in the serum at 2 dpi (Figure S1 A). Minimal virus was found in the heart, which may reflect residual virus from the serum, and virus was not detected in brain at the time of peak lung titer (2 dpi) (Figure S1A). Viral protein was not detected in the heart, liver, small intestine, kidney, or spleen (Figures S1B–S1F). Old mice also exhibited viral titers in the nasal cavity over the first 3 days of infection (peak on 3 dpi at 2 × 104 PFU/tissue), consistent with young adult mice (Figure 3F). Infection with SARS-CoV-2 MA10 also disturbed lung function in aged mice in a similar, but more prolonged, manner compared to young mice with significant changes in PenH, Rpef, and EF50 at 2–5 dpi (Figures 3G–3I).
Figure S1.
SARS-CoV-2 MA10 Does Not Replicate in Non-Respiratory Tract Tissues, Related to Figure 3
Analysis of non-respiratory tract tissues of SARS-CoV-2 MA10 infected 1-year-old female BALB/c mice at 2dpi from Figure 3. (A) Viral titer in serum, heart, and brain. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. (B-F) Representative hematoxylin & eosin (left) and IHC for viral nucleocapsid (right) images of heart (B), liver (C), small intestine (D), kidney (E), and spleen (F). Faint non-specific IHC staining in spleen is result of red blood cell metabolism in macrophages, not viral protein staining. (A, B, E, F) shown at 100X with scale bar representing 500 μm. (D) shown at 200X with scale bar representing 200 μm.
Histological analyses revealed severe DAD and higher ATS ALI scores at later time points throughout the lung in 1-year-old mice (Figures 3J and 3K), consistent with the more pronounced interstitial congestion, epithelial damage, immune cell infiltration, and edema in the older animals (Figure 3L). Viral antigen was detected in small airways and alveolar regions at 2 and 4 dpi (Figure 3L). Viral RNA was also detected in the olfactory epithelium at 2 dpi (Figure 3M), concordant with nasal cavity viral titers (Figure 3F). At 4 dpi, the olfactory epithelium was severely damaged, likely contributing to reduced nasal cavity viral titers, and infection had spread to the Bowman’s gland in the submucosa (Figure 3F).
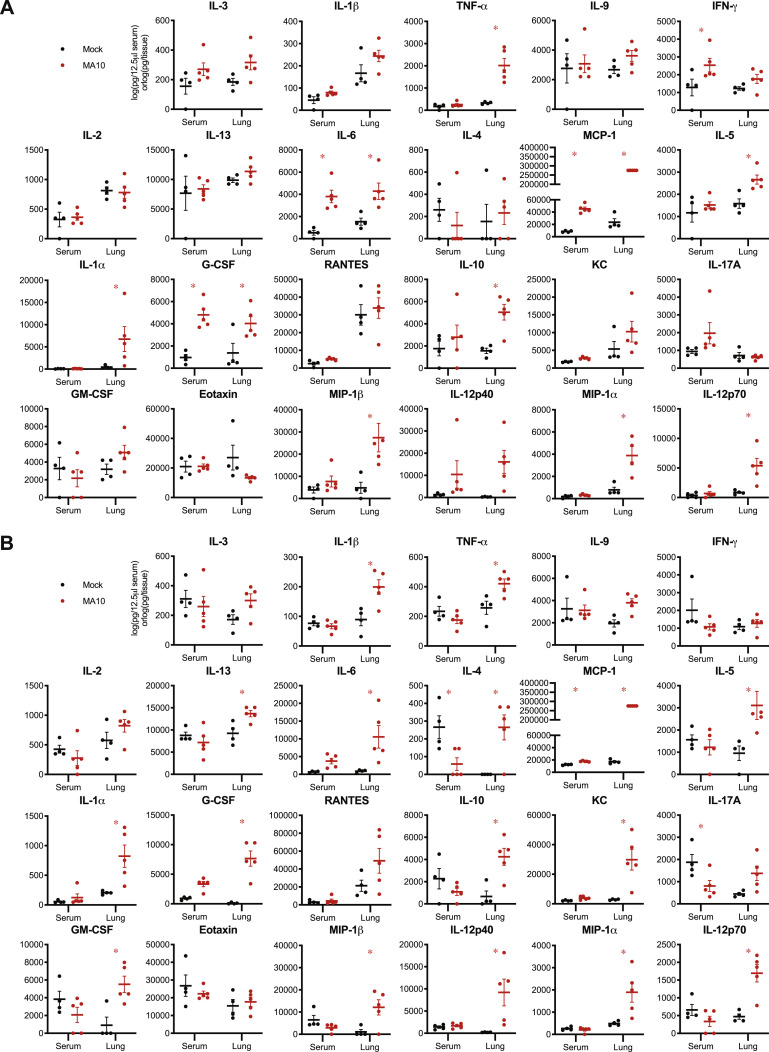
Many viral diseases are associated with a systemic cytokine storm. We analyzed the chemokine and cytokine responses in the serum and lungs of 1-year-old BALB/c mice at 2 and 4 dpi (Figures S2 A and S2B). At 2 dpi, several proinflammatory cytokines were elevated in the lungs of SARS-CoV-2-infected mice, whereas few were elevated systemically in the serum. For instance, IL-6, IL-1α, IL-1β, TNF-α, MCP-1, and IFN-γ were highly elevated in the serum and/or lungs of infected mice, similar to reports in humans (Sinha et al., 2020). It remains uncertain as to whether these elevated cytokines contribute to severe disease outcomes after infection.
Figure S2.
SARS-CoV-2 MA10 Induces Local and Systemic Cytokine and Chemokine Responses, Related to Figure 3
(A-B) Cytokine and chemokine analysis of mock or SARS-CoV-2 MA10 infected 1-year-old BALB/c mice from Figure 3. Serum and lung homogenate were assayed for 23 cytokines and chemokines at 2dpi (A) and 4dpi (B). n = 4 mock and 5 MA10 mice at each time point. Data analyzed by 2-factor ANOVA followed by Sidak’s multiple comparisons. Asterisks represent p < 0.05.
Ameliorated Disease and No Mortality in C57BL/6J Mice after SARS-CoV-2 MA10 Infection
C57BL/6 is the most commonly used mouse strain and is the genetic background for the majority of genetically engineered mice (Bryant, 2011). Because host genetic background dependent differences in disease susceptibility have been described for many infectious diseases including SARS-CoV (Noll et al., 2020; Rasmussen et al., 2014; Gralinski et al., 2015; Graham et al., 2015; Manet et al., 2020), we evaluated SARS-CoV-2 MA10 infection in young adult C57BL/6J mice. In comparison to BALB/c mice, 10-week-old C57BL/6J mice exhibited less severe disease and only the two highest doses (104 and 105 PFU) were associated with significant weight loss, but no mortality after infection (Figure 4 A). Therefore, we performed a detailed analysis of the 104 PFU infectious dose over 7 days for a direct comparison between young BALB/c and C57BL/6J mice. After infection with 104 PFU, C57BL/6J mice exhibited a transient 10%–15% weight loss (peaked on 3 dpi and 4 dpi) (Figure 4B) without mortality (Figure 4C). Gross congestion scores in lungs at the time of harvest never rose above a score of 1 (roughly 25% lung involvement) and declined from 3 dpi until 7 dpi (Figure 4D). A clear peak in viral replication in the lungs was observed on 2 dpi (1.6 × 106 PFU/tissue) that was ∼1 order of magnitude lower than peak titers observed in BALB/c mice. After 2 dpi, titers decreased steadily and were not detectable by 7 dpi (Figure 4E). In addition, viral loads in the nasal cavity were relatively low through 3 dpi (2 × 103 to 1.2 × 104 PFU/tissue) and were undetectable by 4 dpi (Figure 4F). Changes in lung function as measured by WBP were similar in pattern for the two mouse strains, but C57BL/6J mice exhibited restored lung function to near baseline levels by 5 dpi, whereas abnormalities in BALB/c mice persisted until 7 dpi (Figures 4G–4I). Similarly, the patterns of histologic changes were equivalent in C57BL/6J mice to young BALB/c mice (Figure 4L), but the magnitudes of the acute lung injury scores were dramatically attenuated in C57BL/6J mice (Figures 4J and 4K).
Figure 4.
C57BL/6J Mice Display Less Severe Disease following SARS-CoV-2 MA10 Infection
(A) 10-week-old female C57BL/6J mice (n = 5 per group) were mock-infected, infected with 105 PFU SARS-CoV-2 MA, or 102, 103, 104, and 105 PFU SARS-CoV-2 MA10.
(B–L) 10-week-old female BALB/c mice were mock-infected (n = 46) or infected with 104 PFU SARS-CoV-2 MA10 (n = 57).
(B) Percent starting weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons.
(C) Survival rate.
(D) Gross lung congestion score. Data analyzed by 2-factor ANOVA followed by Sidak’s multiple comparisons.
(E) Viral lung titer of mice from (B) (mock-infected: 1 dpi, n = 6; 2 dpi, n = 7; 3 dpi, n = 5; 4 dpi, n = 7; 5 dpi, n = 6; 6 dpi, n = 6; 7 dpi, n = 8; SARS-CoV-2 MA10-infected: 1 dpi, n = 7; 2 dpi, n = 8; 3 dpi, n = 7; 4 dpi, n = 10; 5 dpi, n = 4; 6 dpi, n = 7; 7 dpi, n = 11). Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed as in (D).
(F) Viral nasal cavity titer. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed as in (D).
(G–I) Whole body plethysmography analysis of lung function parameters (10 mice per group at 0 dpi): PenH (G), Rpef (H), and EF50 (I). Data analyzed as in (B).
(J and K) Blinded histopathological evaluation of lung damage using DAD scoring (J) and ATS ALI scoring (K) systems on days 2, 4, and 7 after mock or SARS-CoV-2 MA10 infection. Data analyzed as in (B).
(L) Representative 200× images of lungs from mock and SARS-CoV-2 MA10-infected mice from (J) and (K). H&E shown in the top panels. Bottom panels show immunohistochemistry (IHC) labeling against SARS-CoV-2 nucleocapsid, counterstained with hematoxylin. Scale bars represent 200 μm. Error bars represent SEM about the mean. ∗p < 0.05.
SARS-CoV-2 MA10 Cellular Tropism
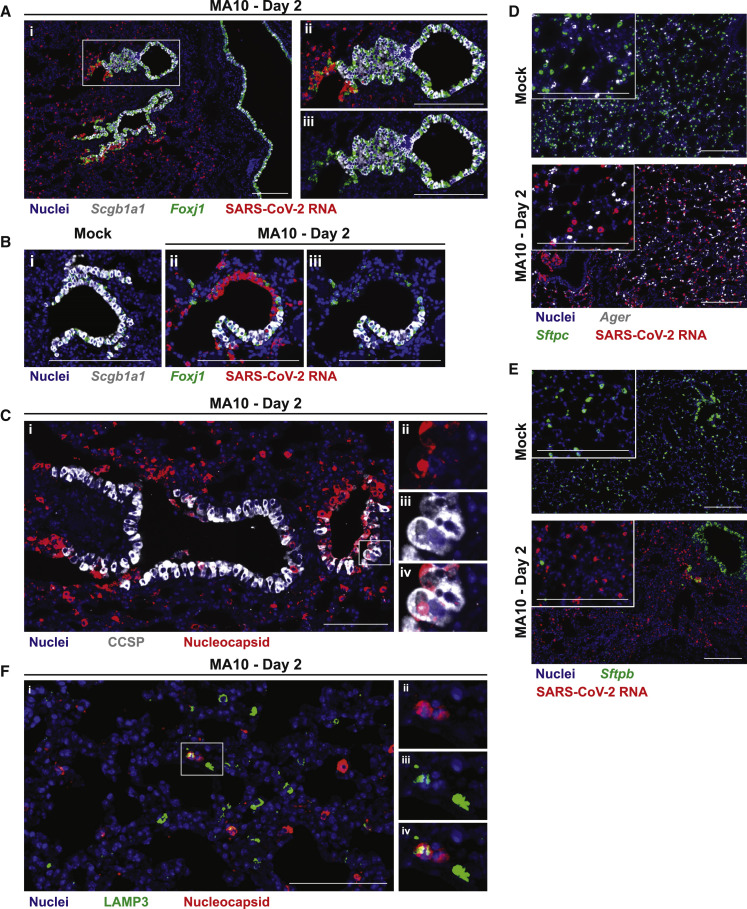
To characterize the tissue and cellular tropism of SARS-CoV-2 MA10, BALB/c mouse nasal and lung tissues were probed for the SARS-CoV-2 viral nucleocapsid and tissue/cell type markers by RNA in situ hybridization (ISH) and immunohistochemistry. In the proximal conducting airways (e.g., trachea, bronchi), little, if any, SARS-CoV-2 MA10 infection was identified. However, robust viral infection was identified in terminal bronchioles that connect to the alveolar spaces (Figures 5A and 5B). In the SARS-CoV-2 MA10-infected mouse terminal bronchiolar epithelial region, expression of Scgb1a1, which is a secretory club cell marker, largely disappeared whereas Foxj1, a ciliated cell marker, persisted. IHC identified SARS-CoV-2 MA10 nucleocapsid expression in non-ciliated cells with occasional colocalization with CCSP (Figures 3L and 5C), suggesting that secretory club cells were infected by SARS-CoV-2 MA10 and subsequently lost Scgb1a1 expression. This cellular tropism of SARS-CoV-2 MA10 is different in human airways, perhaps reflecting different cell levels for ACE2 expression between human versus mice (i.e., ciliated versus secretory club), respectively (Zhang et al., 2020b).
Figure 5.
SARS-CoV-2 MA10 Infects Secretory Club Cells of the Lower Respiratory Tract and Type II Pneumocytes
SARS-CoV-2 MA10 cellular tropism in the lung was assessed by RNA in situ hybridization (ISH) or immunohistochemistry (IHC) in mock or infected female 1-year-old BALB/c mice from Figure 3 at 2 dpi.
(A) ISH of lower airway epithelium. Scgb1a1 is a marker of secretory club cells. Foxj1 is a marker of ciliated cells. (Aii–Aiii) are same inset field with or without SARS-CoV-2 RNA channel.
(B) ISH of terminal bronchiole epithelium. (Bii–Biii) are same field with or without SARS-CoV-2 RNA channel.
(C) IHC of terminal bronchiole epithelium. CCSP is a marker of secretory club cells. (Cii–Civ) are the same inset field as single color channels or merged (Civ).
(D and E) ISH of alveoli. Ager is a marker of type I pneumocytes. Sftpc and Sftpb are markers of type II pneumocytes. (E) IHC of alveoli. LAMP3 is a marker of type II pneumocytes. (Eii–Eiv) are the same inset field as single color channels or merged (Eiv). Scale bars represent 200 μm.
In alveoli, ISH studies of mock-infected mice identified the two major epithelial cell types, i.e., AT1 (Ager expressing) and AT2 (Sftpc, Sftpb expressing) cells (Figures 5D and 5E). In SARS-CoV-2 MA10-infected mice, Sftpc and Sftpb expression characteristic of AT2 cells virtually disappeared, whereas Ager expression associated with AT1 cells persisted at 2 dpi. IHC identified occasional cells expressing a third AT2 cell marker (LAMP3) that also co-expressed the SARS-CoV-2 MA10 nucleocapsid (Figure 3F). Collectively, the loss of surfactant protein transcripts, but not AGER, staining and colocalization of a third AT2 marker (LAMP3) with virus, argues for selective infection by SARS-CoV-2 MA10 of AT2 in the alveolus. The finding that SARS-CoV-2 MA10-infected AT2 cells suppressed expression of selective cell-type-specific genes (e.g., Sftpc and Sftpb) is consistent with findings in infected human AT2 cells in vitro.
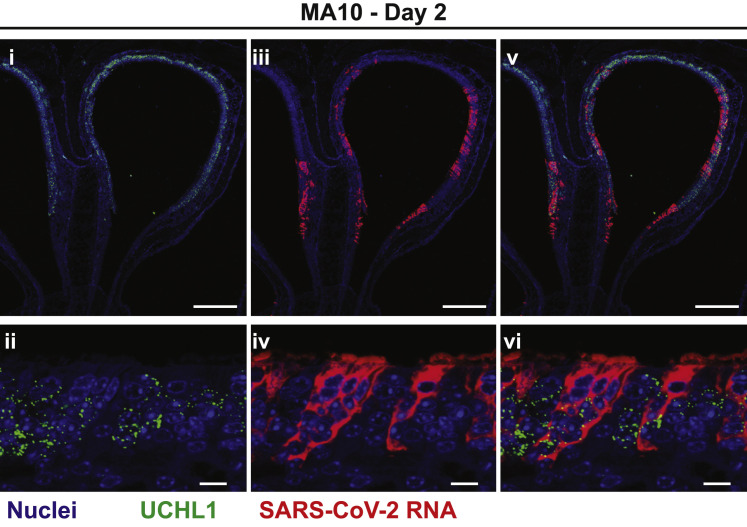
The nasal cavity of the mouse is comprised of ∼50% respiratory epithelium and 50% olfactory epithelium (Chamanza and Wright, 2015). As noted above (Figure 3M), SARS-CoV-2 MA10 RNA was detected in olfactory epithelium, as defined anatomically and by the olfactory sensory neuron marker (OSN) marker, Uchl1 (Figure S3 ). Notably, viral RNA was not detected in cells expressing Uchl1, indicating that SARS-CoV-2 MA10 likely infected sustentacular cells rather than OSNs. The selective olfactory infection by SARS-CoV-2 MA10 is likely associated with altered olfactory function commonly observed in subjects with COVID-19 (Wölfel et al., 2020; Lechien et al., 2020; Spinato et al., 2020).
Figure S3.
SARS-CoV-2 MA10 Infects the Nasal Olfactory Epithelium but Not Olfactory Sensory Neurons, Related to Figure 3
SARS-CoV-2 MA10 cellular tropism in the nasal cavity was assessed by RNA in situ hybridization (ISH) infected female 1-year-old BALB/c mice from Figure 3 at 2dpi. Uchl1 is a marker of olfactory sensory neurons (OSNs). (i & iii and ii and iv) are single color channels shown merged in (v and vi). Scale bars represent 200 μm (i, iii, v) or 10 μm (ii, iv, vi).
Interferon Signaling Is Protective in SARS-CoV-2 MA10 Infection
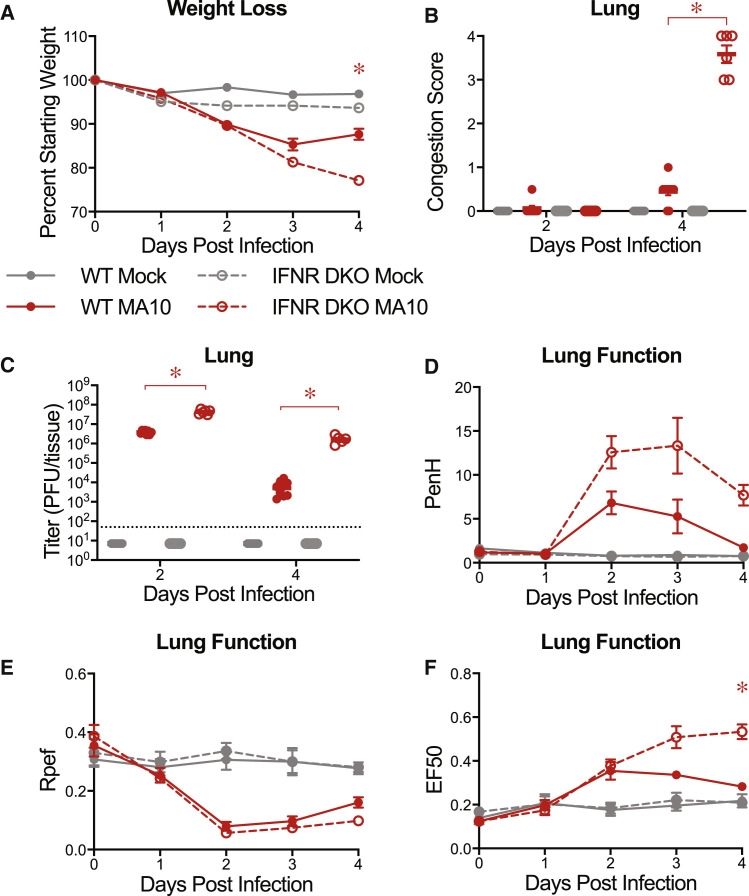
We next tested whether the pathogenic SARS-CoV-2 MA10 virus could be used with genetically deficient mice to elucidate aspects of underlying molecular pathways and networks that regulate SARS-CoV-2 disease. Interferon signaling plays an important role in controlling and regulating disease severity after infection with many viruses, including coronaviruses (Mesev et al., 2019; Israelow et al., 2020). SARS-CoV-2 has also been reported to be sensitive to type I and III interferons in human cells in vitro (Felgenhauer et al., 2020; Vanderheiden et al., 2020) and mice in vivo (Israelow et al., 2020; Dinnon et al., 2020). Consequently, we infected C57BL/6J mice lacking the type I and II interferon receptors (IFNR DKO) and wild-type controls with 104 PFU of SARS-CoV-2 MA10. IFNR DKO mice were more susceptible to SARS-CoV-2 MA10 as indicated by the prolonged weight loss compared to wild-type mice (Figure 6 A). Animals were harvested on planned harvest days. At 4 dpi, SARS-CoV-2 MA10 IFNR DKO mice displayed much higher congestion scores (Figure 6B), which were associated with higher viral titers on 2 and 4 dpi in IFNR DKO mice (Figure 6C). These data suggest that IFNs are important in limiting viral replication and assisting in virus clearance in vivo. Consistent with these data, lung function abnormalities were more pronounced and prolonged in infected IFNR DKO mice (Figures 6C–6E).
Figure 6.
Interferon Signaling Deficient Mice Are More Susceptible to SARS-CoV-2 MA10
10-week-old male and female type I and II interferon receptor double knockout (IFNR DKO; n = 12 mock, n = 19 MA10) and wild-type (WT; n = 11 mock, n = 13 MA10) control mice were mock-infected or infected with 104 PFU SARS-CoV-2 MA10.
(A) Percent starting weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons. Statistical comparisons shown between MA10-infected WT and MA10-infected IFNR DKO mice.
(B) Gross lung congestion score of mice from (A). Data analyzed by 2-factor ANOVA followed by Tukey’s multiple comparisons.
(C) Viral lung titer of mice from (A) (mock-infected: 2 dpi, n = 6 WT and 5 IFNR DKO; 4 dpi, n = 6 WT and 6 IFNR DKO: SARS-CoV-2 MA10-infected: 2 dpi, n = 8 WT and 6 IFNR DKO; 4 dpi, n = 10 WT and 6 IFNR DKO). Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed via 2-factor ANOVA followed by Tukey’s multiple comparisons.
(D–F) Whole body plethysmography analysis of lung function parameters (6 mice per group at 0 dpi): PenH (D), Rpef (E), and EF50 (F). Data analyzed using 2-factor ANOVA followed by Sidak’s multiple comparisons. Error bars represent SEM about the mean. ∗p < 0.05.
SARS-CoV-2 MA10 Allows Rapid Evaluation of Medical Counter Measurements
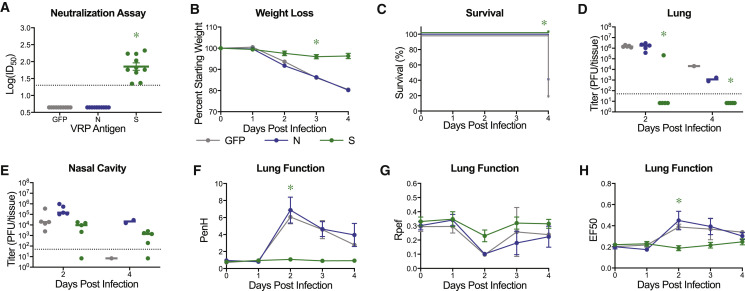
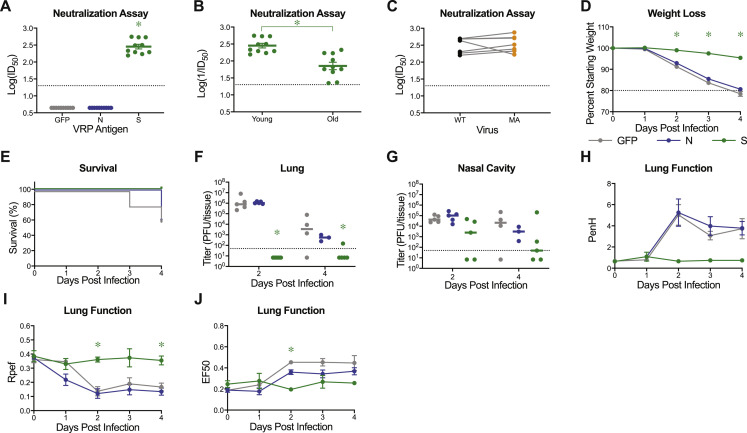
As previously shown for SARS-CoV-2 MA, mouse adapted viral strains allow for rapid testing of prevention and intervention strategies (Dinnon et al., 2020). The SARS-CoV-2 MA10 murine model adds to measurements of viral load the ability to evaluate changes in clinical parameters (weight loss, lung function, and pathologic changes) and mortality (Dinnon et al., 2020). Utilizing our previously described non-select BSL2 Venezuelan equine encephalitis viral replicon particle (VRP) system (Agnihothram et al., 2018; Dinnon et al., 2020), 10-week-old young adult and 1-year-old (“aged”) BALB/c mice were immunized with 103 VRPs expressing SARS-CoV-2 WT spike (S), nucleocapsid (N), and GFP control, followed by a boost at 3 weeks, and challenged with SARS-CoV-2 MA10 4 weeks post boost (7 weeks post prime). Neutralization assays using nLuc expressing reporter virus revealed strongly neutralizing activity in the serum from mice at 3 weeks post boost from spike, but not serum from nucleocapsid or GFP vaccinated mice (Figures 7 A and S4 A). Of note, older (1-year-old) animals exhibited significantly reduced neutralization titers as compared to 10-week-old animals, capturing age related vaccine vulnerabilities often observed in human populations (Figure S4B). Notably, polyclonal sera had similar neutralization titers for both SARS-CoV-2 and SARS-CoV-2 MA containing two of three receptor binding domain changes present in MA10, suggesting that the SARS-CoV-2 MA10 model can accurately be used to test vaccine efficacy (Figure S4C). Only mice vaccinated with SARS-CoV-2 spike expressing VRPs exhibited disease protection as demonstrated by an absence of reductions in in body weight, protection from death (Figures 7B, 7C, S4D, and S4E), and total elimination of viral titers in the lower respiratory tract (lungs) (Figures 7D and S4F). Only one old mouse had detectable lung titer, corresponding to the mouse with the lowest serum neutralization titer. Interestingly, viral titers were still detectable on 2 and 4 dpi after infection in the nasal cavity of immunized young and old mice (Figures 7E and S4G), suggesting that mucosal immunity in the nasal cavity may be difficult to achieve by systemic immunization. Importantly, significant improvements in lung function were measured in both age groups in VRP-S vaccinated mice versus VRP-GFP or VRP-N controls (Figures 7F–7H and S4H–S4J).
Figure 7.
Virus Replicon Particle Delivered Spike Vaccination Protects Old Mice from SARS-CoV-2 MA10 Challenge
1-year-old female BALB/c mice were vaccinated with 103 virus replicon particles (VRPs) expressing SARS-CoV-2 wild-type spike (S, n = 10), nucleocapsid (N, n = 10), or GFP (n = 10). Mice received a boost 3 weeks after prime immunization, and submandibular blood samples were taken to be analyzed via neutralization assays. All mice were challenged 4 weeks after the boost immunization.
(A) Neutralization of SARS-CoV-2 WT by sera from vaccinated mice 3 weeks post boost. ID50, inhibitory concentration necessary to achieve 50% virus neutralization. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed via 1-factor ANOVA followed by Holm-Sidak’s multiple comparisons.
(B) Percent starting weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons.
(C) Survival rate.
(D and E) Viral lung (D) and nasal cavity (E) titer of mice from (B). n = 5 for each group at each time point. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed via 2-factor ANOVA followed by Dunnett’s multiple comparisons.
(F–H) Whole body plethysmography analysis of lung function parameters (n = 5 mice per group at 0dpi): PenH (F), Rpef (G), and EF50 (H). Data analyzed as in (B). Error bars represent SEM about the mean. ∗p < 0.05.
Figure S4.
Virus Replicon Particle Delivered Spike Vaccination Protects Young Mice from SARS-CoV-2 MA10 Challenge, Related to Figure 6
10-week-old female BALB/c mice were vaccinated with 103 virus replicon particles (VRPs) expressing SARS-CoV-2 wild-type spike (S, n = 10), nucleocapsid (N, n = 10), or GFP (n = 10). Mice received a boost 3 weeks after prime immunization and submandibular blood samples were taken to be analyzed via neutralization assays. All mice were challenged 4 weeks after the boost immunization. (A) Neutralization of SARS-CoV-2 WT by sera from vaccinated mice 3 weeks post boost. ID50: inhibitory concentration necessary to achieve 50% virus neutralization. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed via 1-factor ANOVA followed by Holm-Sidak’s multiple comparisons. (B) Comparison of serum IC50 values from 10-week and 1-year-old spike vaccinated mice from Figure 6A. Dotted line represents limit of detection. Data analyzed by unpaired, two-tailed Student’s t test. (C) Comparison of serum IC50 values from 10-week-old spike vaccinated mice to neutralize SARS-CoV-2 WT versus SARS-CoV-2 MA. Data analyzed by Wilcoxon matched-pairs signed rank test. Data not statistically significant. (D) Percent starting weight. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons. (E) Survival rate of mice from (D). (F-G) Viral lung (F) and nasal cavity (G) titer. n = 5 for each group at each time point. Dotted line represents limit of detection. Undetected samples are plotted at half the limit of detection. Log transformed data analyzed via 2-factor ANOVA followed by Dunnett’s multiple comparisons. (G-I) Whole body plethysmography analysis of lung function parameters (n = 5 mice per group at 0dpi): PenH (G), Rpef (H), and EF50 (I). Data analyzed using 2-factor ANOVA followed by Dunnett’s multiple comparisons. Error bars represent standard error of the mean about the mean. Asterisks represent p < 0.05.
Discussion
Mouse models of viral pathogenesis that faithfully recapitulate aspects of human COVID-19 are needed to better understand the underlying molecular mechanisms of disease and assess the performance of medical countermeasures. Herein, we used in vivo experimental evolution to select a mouse-adapted SARS-CoV-2 strain, designated SARS-CoV-2 MA10, capable of causing lethal disease in standard laboratory mice. Importantly, the pathogenic findings of SARS-CoV-2 MA10 increased as a function of mouse age, mirroring age gradients observed in humans (Rothan and Byrareddy, 2020; Li and Ma, 2020). The cellular tropism of SARS-CoV-2 MA10 in the mouse respiratory tract generally reflects that reported in humans (e.g., tropism for AT2 cells and olfactory epithelia) (Hou et al., 2020). Different from humans are the infected secretory (club) cells versus ciliated cells in the conducting airways. The high cytokine expression levels measured in the lungs and to a lesser extent serum of aged animals are consistent with findings reported in humans with ARDS (Song et al., 2020; Costela-Ruiz et al., 2020). Our data also demonstrated that IFN signaling played an important role in attenuating SARS-CoV-2 MA10 viral replication, disease morbidity, and mortality, suggesting that human genetic variation in IFN pathway genes may in part mediate the wide variation of clinical outcomes observed in human SARS-CoV-2 infections. Finally, we provided evidence for the practical application of this model to evaluate SARS-CoV-2 vaccine candidates, with VRP-S immunization protecting and significantly limited viral growth and disease severity in the lung of young and aged mice.
The increased virulence of SARS-CoV-2 MA10 was associated with five mutations acquired through passage in mice. In contrast to SARS-CoV, which acquired nonsynonymous mutations in nsp5, nsp9, nsp13, S, and M when generating mouse adapted SARS-CoV MA15 (Roberts et al., 2007), the SARS-CoV-2 MA10 mouse adaptations reflected amino acid changes in nsp4, nsp7, nsp8, S, and ORF6. Like SARS-CoV MA15, the severity of SARS-CoV-2 MA10 infection was partially attenuated in C57BL/6J mice, providing evidence that host genetic variation in susceptibility and resistance alleles can alter the trajectory of disease and a model of moderate disease. Indeed, a similar intermediate disease phenotype with SARS-CoV MA15 infection in C57BL/6 mice revealed host genes that play protective or pathogenic roles in SARS-CoV disease severity (Gralinski et al., 2013, 2018; Totura et al., 2015; Sheahan et al., 2008; Channappanavar et al., 2016).
With respect to the contribution of the 5 new mutations identified in SARS-CoV-2 MA10, two engineered (Q498Y, P499T) and one evolved (Q493K) amino acid change were noted in the S glycoprotein receptor binding domain, and the latter mutation is predicted to enhance interactions with mouse ACE2 receptor via interaction with residue N31. Other SARS-CoV-2 strains, which replicate but do not produce clinical signs of disease in mice, have RBD mutations at Q498H or R493K, respectively (Wang et al., 2020; Gu et al., 2020). Like for the 2003 SARS-CoV mouse adapted strains (Frieman et al., 2012; Roberts et al., 2007), multiple mutational pathways exist to enhance virus adaptation to the mouse. The mutation in ORF6 is also interesting, because ORF6 acts as an interferon antagonist that blocks nuclear import of karyopherin 2 into the nucleus of the cell (Frieman et al., 2007). Previous studies in our lab demonstrated that deletion of ORF6 attenuated virus pathogenesis, allowing nuclear import of multiple transcription factors and enhanced host defense expression patterns in immortalized human lung cells, Calu3 (Sims et al., 2013). Although speculative, these data suggest that the SARS-CoV-2 MA10 ORF6 mutation may enhance blockade of transcription factor nuclear import, resulting in dampened innate immune antiviral gene expression in the mouse. Finally, mutations were noted in nsp4, nsp7, and nsp8 that have known activities in endoplasmic reticulum and Golgi membrane reorganization to form viral replication factories, as scaffolds for replicase and RNA primase functions, and perhaps processivity activities, respectively (Fehr and Perlman, 2015). Future mapping studies will determine the contribution of each change to viral pathogenesis and host expression patterns in young and aged mice.
Emerging CoVs like SARS-CoV, MERS-Co, and SARS-CoV-2 primarily infect cells lining the upper and lower respiratory tract with damage that triggers the development of ALI, ARDS, and end stage severe lung disease. In humans, many COVID-19 patients exhibit varying degrees of acute injury to airway and alveolar epithelial cells, with resultant fibrin deposition, edema, and hyaline membrane formation. Subsequent hyperplasia of type II pneumocytes, organizing phases of diffuse alveolar damage, focal pulmonary microthrombi, and endothelialitis are also observed in patients (Ackermann et al., 2020; Bradley et al., 2020; Tian et al., 2020). Using established metrics to quantitate pathological features of ALI and ARDS in SARS-CoV (Sheahan et al., 2020b) and MERS-CoV (Sheahan et al., 2020a) mouse models, the lung pathology for SARS-CoV-2 MA10 was quantitated and demonstrated significant ALI and ARDS in an age-related disease gradient. The loss of surfactant protein B and C expression is also consistent with the development of ARDS in SARS-CoV2 MA10-infected mice. Surfactant protein B expression is absolutely required for postnatal lung function, lung compliance, and survival in surfactant protein B knockout mice (Clark et al., 1997; Weaver and Conkright, 2001), whereas lung structure and function in surfactant protein C null mice is normal. Future studies will need to address the real possibility that SARS-CoV-2 MA10 may cause a respiratory distress syndrome (RDS) phenotype primarily associated with surfactant deficiency and whether surfactant replacement therapy might reverse SARS-CoV-2 disease severity when administered early in the viral-dominated phase in mammals (Stevens and Sinkin, 2007; Koumbourlis and Motoyama, 2020). Surfactant protein and RNA expressions are also reduced in lethal SARS-CoV infection in mice, suggesting common mechanisms of respiratory distress across Sarbecoviruses (Gralinski et al., 2013).
The limited availability of transgenic mouse models that can be infected by SARS-CoV-2 has hindered testing of vaccines and therapeutics against this virus. Our data demonstrate that despite the three mutations in the S RBD, alphavirus VRP vaccination with wild-type full-length SARS-CoV-2 S elicited robust neutralization titers against wild-type and SARS-CoV-2 MA10 parental strains. Importantly, these neutralization titers completely protected against SARS-CoV-2 MA10 replication in most mice, which correlated with reduced clinical disease morbidity and mortality. Because aged human populations are most vulnerable to SARS-CoV-2, the use of aged BALB/c or C57BL/6J mice provides a readily available and robust measure of COVID-19 vaccine efficacy. It is noteworthy that several vaccines failed in aged mice challenged with the 2003 mouse-adapted SARS-CoV strain, associated with enhanced Th2 pathology (Sheahan et al., 2011; Tseng et al., 2012). Using alphavirus VRP vectors that drive strong neutralizing antibody responses and Th1 immune responses (Agnihothram et al., 2018), enhanced disease phenotypes were not observed in SARS-CoV-2 MA10-infected aged animals, supporting the importance of vaccines that drive strong Th1 immunity and neutralizing titers to prevent deleterious immune outcomes after vaccination (Bolles et al., 2011; Corbett et al., 2020).
Notably, SARS-CoV-2 replicated efficiently in the nasal cavity of the mice, primarily targeting the olfactory epithelium, where sustentacular and Bowman’s gland cells, but not olfactory neurons, express viral entry components, and support olfactory neuron function (Gupta et al., 2020). Unlike reports in hamsters, replication in mouse olfactory neurons was not evident (Zhang et al., 2020a). Future studies will be needed to elucidate potential relationships between infection of sustentacular cells and chemosensory dysfunction in rodents. Because efficient SARS-CoV-2 transmission may well be associated with efficient high titer replication in the nasal cavity and oral pharynx, the SARS-CoV-2 MA10 model provides an important tool for evaluating vaccine and therapeutic performance in the upper respiratory tract. Our VRP vaccine platform provided limited protection in the nasal cavity of aged mice, suggesting an inability to prevent infection in mucosal as compared to alveolar sites. Because S-IgA antibodies in the secretions of the upper respiratory tract and in saliva appear to result primarily from antigenic stimulation of organized lymphoid follicles of the local mucosa (e.g., pharyngeal, palatine, and lingual tonsils), these data suggest that intranasal vaccination may offer a strategy to protect from upper respiratory SARS-CoV-2 infection (Quiding-Järbrink et al., 1995).
SARS-CoV-2 infection in hamsters results in moderate weight loss and lung pathology (Sia et al., 2020), whereas infections in primates are typically minimally symptomatic (Rockx et al., 2020). Both models provide important metrics for evaluating vaccines and therapeutics and identifying host expression signatures of infection. Although a variety of SARS-CoV-2 mouse models have been reported, these models may have more limited use for studies of alveolar disease pathogenesis. The SARS-CoV-2 MA10 model captures multiple aspects of the COVID-19 syndrome, including a spectrum of morbidity and mortality determined by host genetics and increasing age, and severe pathological features of ALI/ARDS, and corresponding defects in lung function. Accordingly, this model provides the global research community with a robust tool to elucidate the underlying host genetics and molecular mechanisms governing SARS-CoV-2 disease pathogenesis, host expression networks, and immunity after infection. Intermediate disease phenotypes in C57BL/6J mice also provide novel opportunities for using existing mutant mouse resources to determine the role of genes in protective or pathogenic disease outcomes as a function of age. Finally, the capacity to measure vaccine and therapeutic efficacy in high-throughput lethal mouse models of acute lung injury and ARDS may provide critical insights into therapeutic agent performance in the most vulnerable populations (e.g., the elderly and/or in mouse models of the underlying co-morbidities) that contribute to COVID-19 severity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-1 nucleocapsid, dilution at 1:250 | Novus Biologicals | Cat#: NB100-56576; RRID: AB_838838 |
| Rabbit polyclonal SARS coronavirus nucleocapsid, dilution at 1:500 | Invitrogen | Cat#: PA1-41098 RRID: AB_1087200 |
| Goat anti-CCSP, dilution at 1:3000 | Sigma-Aldrich | Cat#: ABS1673 |
| Rat DC-LAMP (LAMP3) antibody, dilution at 1:100 | NOVUS Biologicals | Cat#: DDX0191P-100; RRID: AB_2827532 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555, dilution at 1:1000 | Invitrogen | Cat#: AB_162543; RRID: AB_162543 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, dilution at 1:1000 | Invitrogen | Cat#: AB_2535794; RRID: AB_2535794 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647, dilution at 1:1000 | Thermo Fisher Scientific | Cat#: AB_2535864; RRID: AB_2535864 |
| Discovery OmniMap anti Rabbit HRP | Ventana, Roche | Cat#: 760-4311; RRID: AB_2811043 |
| Bacterial and Virus Strains | ||
| icSARS-CoV-2 WT | Hou et al., 2020 | N/A |
| icSARS-CoV-2 MA | Dinnon et al., 2020 | GenBank: MT844088 |
| SARS-CoV-2 MA10 | This paper | GenBank: MT952602 |
| icSARS-CoV-2-nLuc | Dinnon et al., 2020 | GenBank: MT844089 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRIzol Reagent | ThermoFisher | Cat#: 15596026 |
| Critical Commercial Assays | ||
| Direct-zol RNA Miniprep | Zymo Research | Cat#: R2051 |
| NEBNext Ultra II Non-directional RNA Second Strand Synthesis Module | New England BioLabs | Cat#: E6111S |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Cat#: Q32851 |
| Nextera XT DNA Library Preparation Kits | Illumina | Cat#: FC-131-1096 |
| BioPlex Pro mouse cytokine 23-plex assay | Bio-Rad | Cat#: M60009RDPD |
| Nano-Glo Luciferase Assay | Promega | Cat#: N1130 |
| RNAScope Multiplex Fluorescent Reagent Kit v2 | ACD | Cat#: 323100 |
| RNAScope probe SARS-CoV-2, S gene encoding the spike protein (channel 1, 2, and 3) | ACD | Cat#: 848561 |
| RNAScope probe Ager (channel 1) | ACD | Cat#: 550791 |
| RNAScope probe Sftpc (channel 2) | ACD | Cat#: 314101-C2 |
| RNAScope probe Foxj1 (channel 2) | ACD | Cat#: 317091-C2 |
| RNAScope probe Scgb1a1 (channel 3) | ACD | Cat#: 420351-C3 |
| Vector® TrueVIEW® Autofluorescence Quenching Kit | Vector Laboratories | Cat#: SP-8400 |
| Deposited Data | ||
| Mouse lung total RNA – passage 1 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 2 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 3 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 4 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 5 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 6 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 7 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 8 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 9 | This paper | NCBI Bioproject PRJNA661544 |
| Mouse lung total RNA – passage 10 | This paper | NCBI Bioproject PRJNA661544 |
| Plaque purified cell culture supernatant RNA – Passage 10 plaque 1 | This paper | NCBI Bioproject PRJNA661544 |
| Plaque purified cell culture supernatant RNA – Passage 10 plaque 2 | This paper | NCBI Bioproject PRJNA661544 |
| Plaque purified cell culture supernatant RNA – Passage 10 plaque 3 | This paper | NCBI Bioproject PRJNA661544 |
| Plaque purified cell culture supernatant RNA – Passage 10 plaque 4 | This paper | NCBI Bioproject PRJNA661544 |
| Plaque purified cell culture supernatant RNA – Passage 10 plaque 5 (SARS-CoV-2 MA10) | This paper | NCBI Bioproject PRJNA661544 |
| Experimental Models: Cell Lines | ||
| Simian kidney Vero E6 | ATCC | Cat#: CRL1586; RRID: CVCL_0574 |
| Human bronchiolar airway epithelial cells (HBE) | Marsico Lung Institute (University of North Carolina at Chapel Hill) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: BALB/c: BALB/cAnNHsd | Envigo | Strain 047 |
| Mouse: C57BL/6J | Jackson Laboratory | Strain 000664 |
| Mouse: IFNR DKO | Whitmire Laboratory (University of North Carolina at Chapel Hill) | N/A |
| Oligonucleotides | ||
| Random Primer 9 | New England BioLabs | Cat#: S1254S |
| Software and Algorithms | ||
| CLC Genomics Workbench (Version 12) | QIAGEN | https://digitalinsights.qiagen.com/downloads/product-downloads/ |
| FinePointe (Version 2.3.1.16) | DSI Buxco respiratory solutions, DSI Inc. | https://www.datasci.com/products/software/finepointe-software |
| xPONENT | Luminex | https://www.luminexcorp.com/xponent/ |
| cellSens Entry (Version 3.1) | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| GraphPad Prism (Version 8.4.3) | GraphPad | https://www.graphpad.com |
| Adobe Illustrator (Version 24.2.3) | Adobe | https://www.adobe.com/products/illustrator.html |
| Olyvia (Version 3.1.1) | Olympus | https://olympus-lifescience.com |
| Other | ||
| NanoDropTM One/OneC Microvolume UV-Vis Spectrophotometer | Thermo Fisher Scientific | Cat#: 840-329700 |
| NovaSeq 6000 system | Illumina | https://www.illumina.com/systems/sequencing-platforms/novaseq.html |
| Whole body plethysmography machine | DSI Buxco respiratory solutions, DSI Inc. | https://www.datasci.com/products/buxco-respiratory-products/finepointe-whole-body-plethysmography |
| MAGPIX machine | Luminex | https://www.luminexcorp.com/magpix-system/ |
| Leica ASP 6025 | Leica | https://www.leicabiosystems.com/histology-equipment/tissue-processors/asp6025-s/ |
| Leica Paraplast | Leica | Cat#: 39601006 |
| Ventana Discovery platform | Roche | https://diagnostics.roche.com/us/en/products/instruments/discovery-ultra.html |
| Olympus BX43 light microscope/ Olympus DP27 camera | Olympus | https://www.olympus-lifescience.com/en/microscopes/upright/bx43/?gclid=CjwKCAjw74b7BRA_EiwAF8yHFK_i8yvM2BsQ30kimydPA6qLQlD8f7-lhcGaHpYSdPqsfsvqgpkCKxoC9OsQAvD_BwE |
| SpectraMax M3 luminometer | Molecular Devices | https://www.moleculardevices.com/sites/default/files/en/assets/brochures/br/spectramax-m-series-multi-mode-microplate-readers.pdf |
Resource Availability
Lead Contact
Reagents and resources will be available under material transfer agreements and upon request to the Lead Contact, Ralph S. Baric (rbaric@email.unc.edu).
Material Availability
Material and reagents generated in this study will be made available upon installment of a material transfer agreement (MTA).
Data and Code Availability
Genomic sequence of SARS-CoV-2 MA10, which was generated in this study, has been deposited to GenBank : MT952602). Passaging and plaque purified deep sequencing data have been uploaded to NCBI Bioproject: PRJNA661544.
Experimental Models and Subject Details
Ethics and biosafety
The generation of recombinant SARS-CoV-2 MA and in vivo passaging was approved for use under BSL3 conditions by the University of North Carolina at Chapel Hill Institutional Review Board (UNC-CH IBC) and by a Potential Pandemic Pathogen Care and Oversight committee at the National Institute of Allergy and Infectious Diseases (NIAID). All animal work was approved by Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill according to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the U.S. Department of Agriculture. All work was performed with approved standard operating procedures and safety conditions for SARS-CoV-2. Our institutional BSL3 facilities have been designed to conform to the safety requirements recommended by Biosafety in Microbiological and Biomedical Laboratories (BMBL), the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention (CDC), and the National Institutes of Health (NIH). Laboratory safety plans have been submitted, and the facility has been approved for use by the UNC Department of Environmental Health and Safety (EHS) and the CDC.
Cell lines
Simian kidney cell line Vero E6 (ATTC # CRL1586) was purchased from ATCC and preserved in our laboratory. Primary HBE cell cultures were obtained from the Tissue Procurement and Cell Culture Core Laboratory in the Marsico Lung Institute/Cystic Fibrosis Research Center at UNC. Human tracheobronchial epithelial cells provided by Dr. Scott Randell were obtained from airway specimens resected from patients undergoing surgery under University of North Carolina Institutional Review Board-approved protocols (#03-1396) by the Cystic Fibrosis Center Tissue Culture Core. HBEs were generated by differentiation at an air-liquid interface for 6 to 8 weeks to form well-differentiated, polarized cultures that resembled in vivo pseudostratified mucociliary epithelium (Fulcher et al., 2005).
Virus strains
The virus strains icSARS-CoV-2 WT (Hou et al., 2020), icSARS-CoV-2 MA (Dinnon et al., 2020), and icSARS-CoV-2-nLuc (Dinnon et al., 2020) stocks were generated in our laboratory. SARS-CoV-2 MA10 was generated in this study via serial lung passaging in mice for 10 passages. Briefly, mice were infected with the SARS-CoV-2 MA stock virus (Dinnon et al., 2020) for the first passage and with lung homogenates of the previous passage for all following passages (passage 2 – 10). Clonal isolate from P10 was plaque purified to obtain SARS-CoV-2 MA10. All virus stocks were propagated on Vero E6 cells in minimal essential medium containing 10% fetal bovine serum (HyClone) and supplemented with penicillin/kanamycin (GIBCO). Virus plaques were visualized by neutral red staining for two to three days. All viral infections were conducted under biosafety level 3 (BSL-3) conditions at negative pressure and personnel was protected wearing Tyvek suits connected to personal powered-air purifying respirators.
Mouse strains
10-week and 1-year old female BALB/c/cAnNHsd mice were purchased from Envigo (strain 047), 10-week old female C57BL/6J mice were purchased from Jackson Laboratory (strain 000664), and 10-week old female and male IFNR DKO mice were obtained from the Whitmire Laboratory and bred at the University of North Carolina at Chapel Hill. All animal work was approved by Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill under protocol 19-168 according to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the U.S. Department of Agriculture. All infection studies were performed in animal biosafety level 3 (BSL-3) facilities at University of North Carolina at Chapel Hill.
Method Details
Viruses and cells
The parental SARS-CoV-2 MA virus was derived from an infectious clone of SARS-CoV-2 and further genetically engineered to introduce Q498Y/P499T substitutions into the spike protein (Dinnon et al., 2020). Passage 1 SARS-CoV-2 WT and MA stocks were grown using Vero E6 cells and titered via plaque assay. Briefly, serially diluted virus was added to a monolayer of Vero E6 cells and overlayed with media containing 0.8% agarose. After three days plaques were visualized via staining with Neutral Red dye and counted.
Vero E6 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO), 5% Fetal Clone II serum (Hyclone), and 1X antibiotic/antimycotic (GIBCO). For single step growth curves, cells were infected with a multiplicity of infection (MOI) of 0.5 for 1 hour. After removal of inoculum, cells were washed twice with PBS and 2mL of media added. At designated time points supernatant was harvested and stored at −80°C until further analysis. Well differentiated primary human bronchiolar airway epithelial (HBE) cells were cultured in ALI media. In order to generate a growth curve, cells were infected with a MOI of 0.5 for 2 hours after which the inoculum was removed, cells were rinsed three times with PBS and replaced with media. At designated time points, HAEs were apically washed with 200 μL 1X PBS for 10 minutes and samples stored at −80°C until further analysis.
Clonal isolate from P10 was plaque purified from a plaque assay of a P10-infected mouse lung homogenate via inoculation of Vero E6 cells with an agar stab, generating a passage 1 SARS-CoV-2 MA10 stock. A passage 2 stock was grown, and supernatant viral RNA was sequenced (described below). A larger passage 3 stock was grown, titered, and used for all subsequent experiments.
Sequencing (library preparation and SNP detection)
Viral RNA from clarified cell culture supernatant was isolated using TRIzol LS (Invitrogen) using a Direct-zol RNA Kit (Zymo Research) following manufacturers suggested protocol and quantified by NanoDrop (ThermoFisher Scientific). dsDNA was synthesized by random priming with Random Primer 9 (New England BioLabs) on 500-1000 ng of each isolate’s RNA and reverse transcribed using Super Script II (Sigma-Aldrich) to make cDNA followed by second strand synthesis using NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module (New England BioLabs) following the manufacturer’s suggested protocols. dsDNA was quantified using Qubit dsDNA HS Assay Kit (ThermoFisher Scientific). Libraries were prepared using Nextera XT DNA Library Preparation Kits (Illumina) and sequenced on a NovaSeq 6000 System (Illumina) with paired end reads (2 × 151). SARS-CoV-2 MA10 passage 2 reads were de novo assembled using CLC Genomics Workbench v12 (QIAGEN) to confirm initial viral sequence.
RNA in situ hybridization
RNA-ISH was performed on paraffin-embedded 4 μm tissue sections using the RNAscope Multiplex Fluorescent Assay v2, according to the manufacturer’s instructions (Advanced Cell Diagnostics). Tissue sections were deparaffinized with xylene (2 changes × 5 min) and 100% ethanol (2 changes × 1 min), and then incubated with hydrogen peroxide for 10 min, followed by target retrieval in boiling water for 15 min, and incubation with Protease Plus (Advanced Cell Diagnostics) for 15 min at 40°C. Slides were hybridized with custom probes at 40°C for 2 hours, and signals were amplified according to the manufacturer’s instructions. An Olympus VS200 fluorescent microscope and Olympus confocal microscope were utilized to capture the stained sections.
Immunohistochemistry
Immunohistochemical staining was performed on paraffin-embedded 4 μm tissue sections according to a protocol as previously described (Okuda et al., 2019). Briefly, paraffin-embedded sections were baked at 60 °C for 2–4 hours, and deparaffinized with xylene (2 changes × 5 min) and graded ethanol (100% 2 × 5 min, 95% 1 × 5 min, 70% 1 × 5 min). After rehydration, antigen retrieval was performed by boiling the slides in 0.1 M sodium citrate pH 6.0 (3 cycles with microwave settings: 100% power for 6.5 min, 60% for 6 min, and 60% for 6 min). After cooling and rinsing with distilled water, quenching of endogenous peroxidase was performed with 0.5% hydrogen peroxide in methanol for 15 min, slides washed in PBS, and blocked with 4% normal donkey serum, for an hour at RT. Primary antibodies were diluted in 4% normal donkey serum in PBST and incubated over night at 4 °C. Species-matched gamma globulin was used as an isotype control at the same concentration as the primary antibody. Sections were washed in PBST and Species-matched secondary antibodies were applied for 60 min at RT. After washing in PBST, the Vector® TrueVIEW Autofluorescence Quenching Kit (Vector laboratories) was used to reduce background staining, and glass coverslips were placed over tissue sections with the ProLong Gold Antifade Reagent with DAPI (Invitrogen). Coverslipped slides were scanned and digitized using an Olympus VS200 whole slide scanner microscope.
In vivo infection
BALB/cAnNHsd mice were obtained from Envigo (strain 047). C57BL/6J mice were obtained from the Jackson Laboratory (strain 000664). Type I and II interferon receptor double knock out (IFNR DKO) mice were originally obtained from the Whitmire laboratory and bred at the University of North Carolina at Chapel Hill. Anesthetized (ketamine/xylazine) mice were intranasally infected with 105 PFU SARS-CoV-2 MA and different doses of SARS-CoV-2 MA10 diluted in PBS where indicated. Clinical signs of disease (weight loss and lung function) were monitored daily. Lung function was assessed utilizing whole body plethysmography (WBP; DSI Buxco respiratory solutions, DSI Inc.) by allowing mice to acclimate in WBP chambers for 30 minutes followed by 5 minutes of data recording as described previously (Menachery et al., 2015). Acquired data was analyzed using FinePointe software. Mice were euthanized by isoflurane overdose at indicated time points when samples for titer (caudal right lung lobe) and histopathological analyses (left lung lobe) were collected. All animals in this manuscript that are recorded as “dead” were either found dead in cage or were moribund and euthanized as they approached 70% of their starting body weight which is the defined human endpoint according to the respective animal protocol. Importantly, mice were randomized and assigned to specific harvest days before the start of the experiment. Lung viral titers were determined by plaque assay. Briefly, right caudal lung lobes were homogenized in 1mL PBS using glass beads and serial dilutions of the clarified lung homogenates were added to a monolayer of Vero E6 cells. After three days plaques were visualized via staining with Neutral Red dye and counted. The left lung lobe was stored in 10% phosphate buffered formalin for 7 days prior to removal from the BSL3 for processing. After paraffin embedding, sectioning and staining histopathological scoring was performed.
Chemokine & Cytokine analysis
BioPlex Pro mouse cytokine 23-plex assay (Bio-Rad) was utilized to analyze chemokines and cytokines in serum and lung samples from 1-year-old BALB/c mice according to manufacturer’s protocol. 50 μL of clarified lung samples or 50 μL of 1:4 diluted serum samples were incubated with magnetic capture beads, washed, incubated with detection antibodies and SA-PE. Cytokines were recorded on a MAGPIX machine (Luminex) and quantitated via comparison to a standard curve. xPONENT software was used for data collection and analysis.
Histological analysis and antigen staining
Immediately after euthanasia, the left lung lobe was harvested and fixed by submersion in 10% phosphate buffered formalin for 7 days. Fixed tissues were routinely processed on a Leica ASP 6025, embedded in paraffin (Leica Paraplast) and sectioned at 4um thickness. Sequential sections were stained with hematoxylin and eosin (Richard Allan Scientific) and stained for SARS-CoV-2 nucleocapsid using a monoclonal anti-SARS-CoV nucleocapsid antibody (NB100-56576, Novus Biologicals) on the Ventana Discovery platform (Roche). Briefly, antigen retrieval was performed using Ventana’s CC1 (pH 8.5), tissues were blocked, primary antibody diluted at 1:250 using Discovery Casein Diluent (760-219, Roche), ready-to use secondary antibody (Discovery OmniMap anti Rabbit HRP, 760-4311), followed by DAB development and Hematoxylin II staining. Pathology was evaluated and scored by a board-certified veterinary pathologist (S.A.M). Lung histopathology was blindly scored using three 600X fields per tissue as previously described. Briefly, three random fields of diseased tissue at 600X total magnification (60X objective x 10X eyepiece) were chosen and scored in a blinded manner for ALI and DAD histological scoring systems. For the ASL/ALI scoring system the following parameter were analyzed: (A) neutrophils in the alveolar space (none = 0, 1-5 cells = 1, > 5 cells = 2); (B) neutrophils in the interstitial space/septae (none = 0, 1-5 cells = 1, > 5 cells = 2); (C) hyaline membranes (none = 0, one membrane = 1; > 1 membrane = 2); (D) proteinaceous debris in air spaces (none = 0, one instance = 1, > 1 instance = 2); (E) alveolar septal thickening (> 2x mock thickness = 0, 2-4x mock thickness = 1, > 4x mock thickness = 2). Scores were calculated as followed: [(20 x A) + (14 x B) + (7 x C) + (7 x D) + (2 x E)] / 100. Final scores were obtained by averaging three fields per mouse. The defuse alveolar damage (DAD) scores were determined as followed: 1 = absence of cellular sloughing and necrosis; 2 = uncommon solitary cell sloughing and necrosis; 3 = multifocal (3 + foci) cellular sloughing and necrosis with uncommon septal wall hyalinization; 4 = multifocal (> 75% of field) cellular sloughing and necrosis with common and/or prominent hyaline membranes. The average of three fields determined the final DAD score per mouse. An Olympus BX43 light microscope was used to capture images at 200X magnification with a DP27 camera using cellSens Dimension software.
Vaccination and neutralization studies
Mice were vaccinated with Venezuelan equine encephalitis virus strain 3526 based replicon particles (VRPs) expressing SARS-CoV-2 spike (S), nucleocapsid (N), or GFP as control. VRPs were given via hind footpad injection at a dose of 103 in 10 μL. The same strategy was used to boost mice 3 weeks post prime and presence of neutralizing antibodies was confirmed in submandibular bleeds at the time of boost.
Authentic virus neutralization of sera from 3 weeks post boost using nanoLuciferase-expressing SARS-CoV-2 virus (SARS-CoV-2 nLuc), bearing wild-type spike protein, was performed as described with slight modification (Hou et al., 2020; Dinnon et al., 2020). Briefly, Vero E6 cells were seeded at 2x104 cells/well in a 96-well plate 24h before the assay. 100 PFU of SARS-CoV-2-nLuc virus were mixed with serial diluted sera at 1:1 ratio and incubated at 37°C for 1h. A 8-point, 3-fold dilution curve was generated for each sample with starting concentration at 1:20. Virus and Ab mix was added to cells and incubated at 37°C + 5% CO2 for 48h. Luciferase activities were measured by Nano-Glo Luciferase Assay System (Promega, WI) following manufacturer protocol using SpectraMax M3 luminometer (Molecular Device). Percent inhibition and 50% inhibition concentration (IC50) were calculated by the following equation: [1-(RLU with sample/ RLU with mock treatment)] x 100%. Fifty percent inhibition titer (IC50) was calculated in GraphPad Prism 8.4.2 by fitting the data points using a sigmoidal dose-response (variable slope) curve.
Intranasal challenge of anesthetized (ketamine/xylazine) mice with 104 PFU SARS-CoV-2 MA10 was performed 4 weeks post boost. Changes in body weight and alterations in lung function parameters were recorded daily and mice were euthanized by isoflurane overdose for harvests on day 2 and day 4 after infection. Viral titers in lungs were analyzed via plaque assay.
Quantification and Statistical Analysis
Data visualization and analyses were performed using build-in functions of GraphPad Prism. Specific statistical tests, numbers of animals, and definitions of center, dispersion and precision measures are mentioned in respective figure legends. For the characterization of SARS-CoV-2 MA10 in young and old BALB/c as well as young C57BL/6 mice the following statistical tests were used: mixed effect analysis followed by Sidak’s multiple comparisons was used to analyze weight loss and whole body plethysmography data; cell growth curves and cytokine / chemokine responses were analyzed by 2-factor ANOVA followed by Sidak’s multiple correction; gross lung congestions scores, lung and nasal titers, as well as DAD and ATS / ALI scores were analyzed by 2-factor ANOVA followed by Sidak’s multiple comparisons; survival rates were analyzed by log rank test. For the IFNR-DKO data the following statistical tests were used: weight loss data was analyzed using mixed effect analysis followed by Sidak’s multiple comparisons; 2-factor ANOVA followed by Tukey’s multiple comparisons was used for gross congestion scores and lung / nasal titer data; whole body plethysmography was analyzed via 2-factor ANOVA followed by Sidak’s multiple comparisons. VRP mouse data was analyzed as followed: weight loss and whole body plethysmography data was analyzed via mixed effect analysis followed by Sidak’s multiple comparisons; neutralization data was log transformed and analyzed via 1-factor ANOVA followed by Holm-Sidak’s multiple comparison; lung / nasal titer data as well as whole body plethysmography was analyzed by 2-factor ANOVA followed by Dunnetts multiple comparisons; unpaired, two-tailed Student’s t test was used for comparisons of serum IC50 values from 10-week and 1-year-old vaccinated mice; Comparison of serum IC50 values from 10-week-old spike vaccinated mice to neutralize SARS-CoV-2 WT versus SARS-CoV-2 MA was analyzed via Wilcoxon matched-pairs signed rank test.
Acknowledgments
This project was supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly. Animal histopathology service was performed by Dawud Hilliard, Lily Wai, Ling Wang, and Mia Evangelista in the Animal Histopathology and Laboratory Medicine Core at the University of North Carolina, which is supported in part by an NCI Center Core Support Grant (5P30CA016086-41) to the UNC Lineberger Comprehensive Cancer Center. This project was funded in part by the National Institute of Allergy and Infectious Diseases, NIH, U.S. Department of Health and Human Services award 1U19 AI142759 (Antiviral Drug Discovery and Development Center awarded to R.S.B), 5R01AI132178 (partnership grant awarded to T.P.S. and R.S.B), AI100625, and AI108197 to R.S.B. as well as an animal models contract from the NIH (HHSN272201700036I 75N93020F00001) and U54CA260543 sponsored by NCI. K.H.D. was funded by T32AI142759.
Author Contributions
S.R.L. and K.H.D. designed and conducted in vitro and animal experiments, analyzed data, generated figures, and wrote the manuscript. A.S. conducted in vitro and animal experiments and edited the manuscript. L.V.T. conducted in vitro experiments and neutralization assays and analyzed data. C.E.E. generated VRP vaccine and conducted in vitro experiments. K.O. conducted in vitro experiments and imaged and analyzed data. Y.J.H. and E.J.F. conducted in vitro experiments. W.S. performed and analyzed sequencing. S.A.M. imaged and analyzed histology. A.W., K.L.G., and A.J.B. assisted with animal experiments. T.S. assisted with in vitro experiments. T.P.S. wrote manuscript. L.E.G. conducted animal experiments and edited manuscript. N.J.M. analyzed data. R.C.B. analyzed data and edited the manuscript. R.S.B. supervised the project, assisted in experimental design, and wrote the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Published: September 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.050.
Supplemental Information
References
- Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnihothram S., Menachery V.D., Yount B.L., Jr., Lindesmith L.C., Scobey T., Whitmore A., Schäfer A., Heise M.T., Baric R.S. Development of a Broadly Accessible Venezuelan Equine Encephalitis Virus Replicon Particle Vaccine Platform. J. Virol. 2018;92:e00027-18. doi: 10.1128/JVI.00027-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C.D. The blessings and curses of C57BL/6 substrains in mouse genetic studies. Ann. N Y Acad. Sci. 2011;1245:31–33. doi: 10.1111/j.1749-6632.2011.06325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N., Pewe L., Trandem K., Perlman S. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology. 2006;347:410–421. doi: 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamanza R., Wright J.A. A Review of the Comparative Anatomy, Histology, Physiology and Pathology of the Nasal Cavity of Rats, Mice, Dogs and Non-human Primates. Relevance to Inhalation Toxicology and Human Health Risk Assessment. J. Comp. Pathol. 2015;153:287–314. doi: 10.1016/j.jcpa.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E.W., Zachariah P., Gorelik M., Boneparth A., Kernie S.G., Orange J.S., Milner J.D. Multisystem Inflammatory Syndrome Related to COVID-19 in Previously Healthy Children and Adolescents in New York City. JAMA. 2020;324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.C., Weaver T.E., Iwamoto H.S., Ikegami M., Jobe A.H., Hull W.M., Whitsett J.A. Decreased lung compliance and air trapping in heterozygous SP-B-deficient mice. Am. J. Respir. Cell Mol. Biol. 1997;16:46–52. doi: 10.1165/ajrcmb.16.1.8998078. [DOI] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020 doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgenhauer U., Schoen A., Gad H.H., Hartmann R., Schaubmar A.R., Failing K., Drosten C., Weber F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020 doi: 10.1074/jbc.AC120.013788. Published online June 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., Randell S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- Graham J.B., Thomas S., Swarts J., McMillan A.A., Ferris M.T., Suthar M.S., Treuting P.M., Ireton R., Gale M., Jr., Lund J.M. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6 doi: 10.1128/mBio.00493-15. e00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Bankhead A., 3rd, Jeng S., Menachery V.D., Proll S., Belisle S.E., Matzke M., Webb-Robertson B.J., Luna M.L., Shukla A.K., et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. MBio. 2013;4 doi: 10.1128/mBio.00271-13. e00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J., et al. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Sheahan T.P., Morrison T.E., Menachery V.D., Jensen K., Leist S.R., Whitmore A., Heise M.T., Baric R.S. Complement Activation Contributes to Severe Acute Respiratory Syndrome Coronavirus Pathogenesis. MBio. 2018;9:e01753-18. doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020 doi: 10.1126/science.abc4730. Published online July 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Mohanty S.K., Mittal A., Kalra S., Kumar S., Mishra T., Ahuja J., Sengupta D., Ahuja G. The Cellular basis of the loss of smell in 2019-nCoV-infected individuals. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa168. Published online August 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T, Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201241. e20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumbourlis A.C., Motoyama E.K. Lung Mechanics in COVID-19 Resemble Respiratory Distress Syndrome, Not Acute Respiratory Distress Syndrome: Could Surfactant Be a Treatment? Am. J. Respir. Crit. Care Med. 2020;202:624–626. doi: 10.1164/rccm.202004-1471LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020;277:2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit. Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manet C., Simon-Lorière E., Jouvion G., Hardy D., Prot M., Conquet L., Flamand M., Panthier J.J., Sakuntabhai A., Montagutelli X. Genetic Diversity of Collaborative Cross Mice Controls Viral Replication, Clinical Severity, and Brain Pathology Induced by Zika Virus Infection, Independently of Oas1b. J. Virol. 2020;94 doi: 10.1128/JVI.01034-19. e01034-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R., Qiu Y., He J.S., Tan J.Y., Li X.H., Liang J., Shen J., Zhu L.R., Chen Y., Iacucci M., et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G., Downey G., Moore B.B., Groshong S.D., Matthay M.A., Slutsky A.S., Kuebler W.M., Acute Lung Injury in Animals Study Group An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New Metrics for Evaluating Viral Respiratory Pathogenesis. PLoS ONE. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll K.E., Whitmore A.C., West A., McCarthy M.K., Morrison C.R., Plante K.S., Hampton B.K., Kollmus H., Pilzner C., Leist S.R., et al. Complex Genetic Architecture Underlies Regulation of Influenza-A-Virus-Specific Antibody Responses in the Collaborative Cross. Cell Rep. 2020;31:107587. doi: 10.1016/j.celrep.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Chen G., Subramani D.B., Wolf M., Gilmore R.C., Kato T., Radicioni G., Kesimer M., Chua M., Dang H., et al. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am. J. Respir. Crit. Care Med. 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiding-Järbrink M., Granström G., Nordström I., Holmgren J., Czerkinsky C. Induction of compartmentalized B-cell responses in human tonsils. Infect. Immun. 1995;63:853–857. doi: 10.1128/iai.63.3.853-857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R., et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.E., Knudson C.J., Hartwig S.M., Pewe L.L., Meyerholz D.K., Langlois R.A., Harty J.T., Varga S.M. Memory CD8 T cells mediate severe immunopathology following respiratory syncytial virus infection. PLoS Pathog. 2018;14:e1006810. doi: 10.1371/journal.ppat.1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4:e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Whitmore A., Long K., Ferris M., Rockx B., Funkhouser W., Donaldson E., Gralinski L., Collier M., Heise M., et al. Successful vaccination strategies that protect aged mice from lethal challenge from influenza virus and heterologous severe acute respiratory syndrome coronavirus. J. Virol. 2011;85:217–230. doi: 10.1128/JVI.01805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schäfer A., Matzke M.M., Webb-Robertson B.J., Chang J., Luna M.L., Long C.E., et al. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P., Matthay M.A., Calfee C.S. Is a “Cytokine Storm” Relevant to COVID-19? JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.3313. Published online June 30, 2020. [DOI] [PubMed] [Google Scholar]
- Song P., Li W., Xie J., Hou Y., You C. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinato G., Fabbris C., Polesel J., Cazzador D., Borsetto D., Hopkins C., Boscolo-Rizzo P. Alterations in Smell or Taste in Mildly Symptomatic Outpatients With SARS-CoV-2 Infection. JAMA. 2020;323:2089–2090. doi: 10.1001/jama.2020.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T.P., Sinkin R.A. Surfactant replacement therapy. Chest. 2007;131:1577–1582. doi: 10.1378/chest.06-2371. [DOI] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J., et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., Liu S.S., Zhang N.N., Li X.F., Xiong R., et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020;28:124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Xiong Y., Liu H., Niu L., Guo J., Liao M., Xiao S.Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod. Pathol. 2020;33:1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. MBio. 2015;6:e00638-15. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. Type I and Type III IFN Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020;94:e00985-20. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shuai L., Wang C., Liu R., He X., Zhang X., Sun Z., Shan D., Ge J., Wang X., et al. Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell. 2020 doi: 10.1007/s13238-020-00767-x. Published online August 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver T.E., Conkright J.J. Function of surfactant proteins B and C. Annu. Rev. Physiol. 2001;63:555–578. doi: 10.1146/annurev.physiol.63.1.555. [DOI] [PubMed] [Google Scholar]
- Wichmann D., Sperhake J.P., Lütgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schröder A.S., et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zhang A.J., Lee A.C., Chu H., Chan J.F., Fan Z., Li C., Liu F., Chen Y., Yuan S., Poon V.K., et al. SARS-CoV-2 infects and damages the mature and immature olfactory sensory neurons of hamsters. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa995. Published online July 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rostami M.R., Leopold P.L., Mezey J.G., O’Beirne S.L., Strulovici-Barel Y., Crystal R.G. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am. J. Respir. Crit. Care Med. 2020;202:219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequence of SARS-CoV-2 MA10, which was generated in this study, has been deposited to GenBank : MT952602). Passaging and plaque purified deep sequencing data have been uploaded to NCBI Bioproject: PRJNA661544.