Abstract
Novel COVID-19 therapeutics are urgently needed. We generated a phage-displayed human antibody VH domain library from which we identified a high-affinity VH binder ab8. Bivalent VH, VH-Fc ab8, bound with high avidity to membrane-associated S glycoprotein and to mutants found in patients. It potently neutralized mouse-adapted SARS-CoV-2 in wild-type mice at a dose as low as 2 mg/kg and exhibited high prophylactic and therapeutic efficacy in a hamster model of SARS-CoV-2 infection, possibly enhanced by its relatively small size. Electron microscopy combined with scanning mutagenesis identified ab8 interactions with all three S protomers and showed how ab8 neutralized the virus by directly interfering with ACE2 binding. VH-Fc ab8 did not aggregate and did not bind to 5,300 human membrane-associated proteins. The potent neutralization activity of VH-Fc ab8 combined with good developability properties and cross-reactivity to SARS-CoV-2 mutants provide a strong rationale for its evaluation as a COVID-19 therapeutic.
Keywords: human VH antibody domain, virus neutralization, electron microscopy, SARS-CoV-2, mouse and hamster models
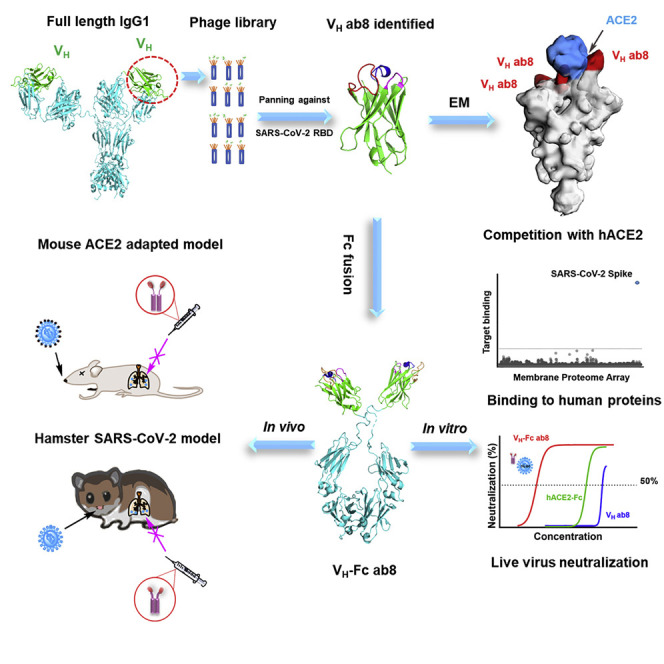
Graphical Abstract
Highlights
- •
A high-affinity human antibody domain, VH ab8, specific for SARS-CoV-2 was selected
- •
VH ab8 bound to all three S protomers competing with ACE2
- •
Bivalent VH, VH-Fc ab8, potently neutralized SARS-CoV-2 in vitro and in animals
- •
Small size and bivalency contribute to the high ab8 SARS-CoV-2 neutralizing potency
A high-affinity human antibody domain, VH ab8, specific for SARS-CoV-2, bound to all three S protomers competing with ACE2. The relatively small size and bivalency of VH-Fc ab8 contributed to its high potency in two animal models of infection.
Introduction
The global outbreak of a severe acute respiratory distress (SARS) coronavirus 2 (SARS-CoV-2) associated disease 2019 (COVID-19) requires rapid identification of therapeutics and vaccines. While many vaccines are in clinical development, the time to market can be relatively long, and immunogenicity can be limited for high-risk groups (Amanat and Krammer, 2020). Alternatively and complementarily, antibodies can be used as safe and effective prophylactics and therapeutics (Pelegrin et al., 2015). Convalescent plasma from COVID-19 patients inhibited SARS-CoV-2 infection and alleviated symptoms of newly infected patients (Casadevall and Pirofski, 2020; Rojas et al., 2020) suggesting that potent neutralizing monoclonal antibodies (mAbs) may be even more effective.
SARS-CoV-2 genome shares more than 80% homology to the SARS-CoV (Li et al., 2020b). Similar to SARS-CoV, SARS-CoV-2 uses the spike (S) envelope glycoprotein to enter into host cells. The viral entry is initiated by the receptor binding domain (RBD) of the S protein binding to its receptor, angiotensin-converting enzyme 2 (ACE2), leading to conformational change of the S2 subunit and formation of six helical-bundle resulting in membrane fusion between viral and host cells (Jiang et al., 2020; Yan et al., 2020). The SARS-CoV RBD contains immune-dominant epitopes that can elicit neutralizing antibodies conferring protection to SARS-CoV infection (He et al., 2005). A recent bioinformatics study showed that SARS-CoV-2 RBD has several B cell epitopes (Grifoni et al., 2020). SARS-CoV-2 RBD-based immunogens were able to elicit neutralizing sera in animals (Quinlan et al., 2020). Thus, SARS-CoV-2 RBD is a good target for developing potent neutralizing mAbs. We and others have identified such potent neutralizing human mAbs targeting the RBD of SARS-CoV (Zhu et al., 2007) and the middle east respiratory syndrome coronavirus (MERS-CoV) (Ying et al., 2014a). Recently, several groups have reported the isolation of potent neutralizing antibodies from convalescent human donors but all are in an immunoglobulin G1 (IgG1) format with a molecular mass of ~150 kDa (Cao et al., 2020; Ju et al., 2020; Rogers et al., 2020; Shi et al., 2020; Zost et al., 2020).
Antibody domains and fragments such as Fab (fragment antigen binding, molecular weight of 50 kDa), scFv (singe-chain variable fragment, 30 kDa), and VH (heavy chain variable domain, 15 kDa) are attractive antibody formats as candidate therapeutics (Nelson, 2010). For example, isotope-labeled antibody fragments are more suitable for bio-imaging due to their better tissue penetration and faster clearance compared to full-size antibodies (Freise and Wu, 2015). Single antibody domains (sAbd; e.g., camelid VHH [15 kDa]) exhibit strong antigen binding and high stability (Harmsen and De Haard, 2007). We and others have demonstrated that human IgG1 heavy chain variable domain (VH) can be engineered to achieve high stability and affinity to antigens (Nilvebrant et al., 2016), as exemplified by the VH, m36.4, targeting the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein co-receptor binding site (Chen et al., 2008a). The VH domains small size could improve therapeutic efficacy for infectious diseases, such as COVID-19 because of greater penetration to sites of infection. The conformation of the SARS-CoV-2 S trimer is dynamic with only one RBD in the “up” conformation presenting neutralizing epitopes while epitopes in the other two RBDs may be masked (Yan et al., 2020). Small VHs may achieve binding to the cryptic RBD epitopes during the dynamic “breathing” of the S trimer (Liu et al., 2020). In addition, VHs may have an advantage for treatment of respiratory virus infections, because VHs could efficiently penetrate tissue, especially when using direct delivery through inhalation (Detalle et al., 2015).
To identify potent neutralizing VHs against SARS-CoV-2, we panned our large (1011 clones) and diverse phage-displayed human VH antibody library against recombinant RBD. Several VH binders were isolated and screened for their affinities, ACE2 competition, and stabilities. One of those VHs, ab8, in an Fc (human IgG1, crystallizable fragment) fusion format, showed potent neutralization activity and specificity against SARS-CoV-2 both in vitro and in two animal models. To our knowledge, this is the first report for high potency of a human antibody domain (VH) in two animal models of infection.
Results
Selection of a High-Affinity VH, ab8, and Its Conversion to a VH-Fc
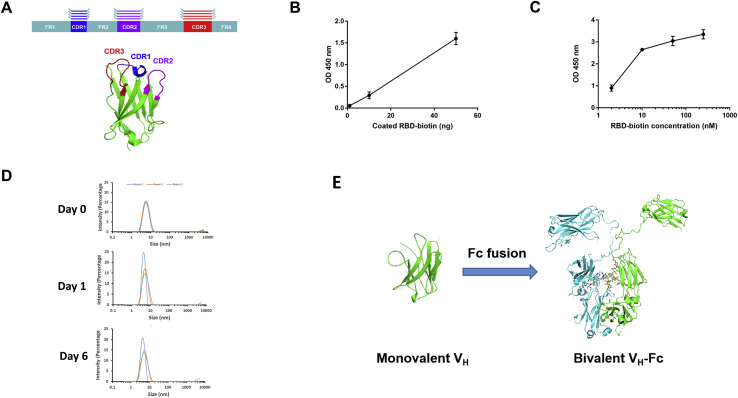
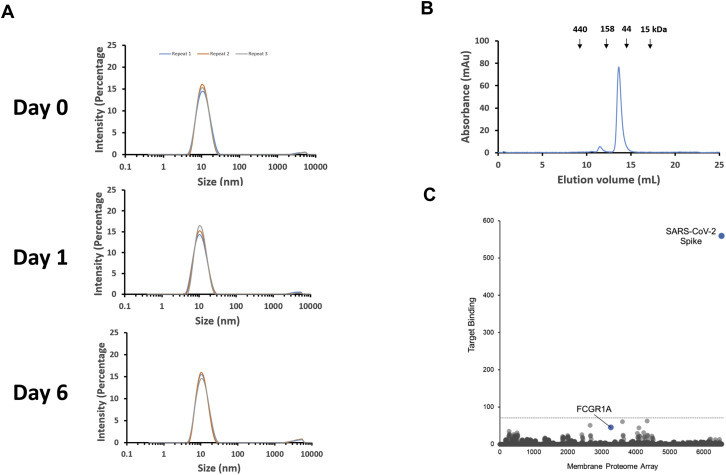
We generated a large phage-displayed human VH library where heavy chain complementarity-determining regions (HCDR1, HCDR2, HCDR3 s) were grafted into their cognate positions of a stable scaffold based on the germline VH3-23 (Figure S1 A). It was panned against recombinant RBD antigens with two different tags (avi-his and human IgG1 Fc tag) that were sequentially used to avoid phage enrichment to tags and related epitopes. The quality of the RBD used for panning was confirmed by ACE2 binding (Figures S1B and S1C). After three rounds of panning, a panel of VH binders was obtained. Among the highest affinity binders, we selected one, VH ab8, which did not aggregate during a 6-day incubation at 37°C as tested by dynamic light scattering (DLS) (Figure S1D). To increase the VH ab8 avidity and extend its in vivo half-life, it was converted to a bivalent antibody domain by fusion to the human IgG1 Fc (VH-Fc ab8) (Figure S1E).
Figure S1.
Schematic Representation of VH Library Construction Strategy, Characterization of the RBD-His-Biotin as an Antigen for Panning, and Evaluation the Aggregation Propensity of VH ab8, Related to STAR Methods
(A) Schematic representation of HCDRs grafting into their cognate positions on a stable scaffold. (B) ELISA of biotinylated RBD330-532 binding to streptavidin-HRP. (C) ELISA measurement of binding of biotinylated RBD-his to ACE2. ~100 ng ACE2-Fc was coated on plate with incubation of serially diluted RBD-his-biotin. Binding was detected by using HRP conjugated streptavidin. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. (D) Evaluation of aggregation of VH ab8 by DLS. VH ab8 (4 mg/ml) in PBS was incubated at 37°C. On day 0, day 1 and day 6, samples were taken out for DLS measurement. All measurements were repeated by three times. (F) Scheme of conversion of VH ab8 into VH-Fc ab8 by fusing IgG1 Fc. The linker between VH and Fc is the natural human IgG1 upper and lower hinge (DKTHTCPPCPAPELL). VH ab8 and VH-Fc ab8 structure is modeled by the online SWISS-MODEL sever (https://swissmodel.expasy.org/).
High-Avidity-Specific Binding of VH-Fc ab8 to RBD and Cell Surface-Associated Native S Protein
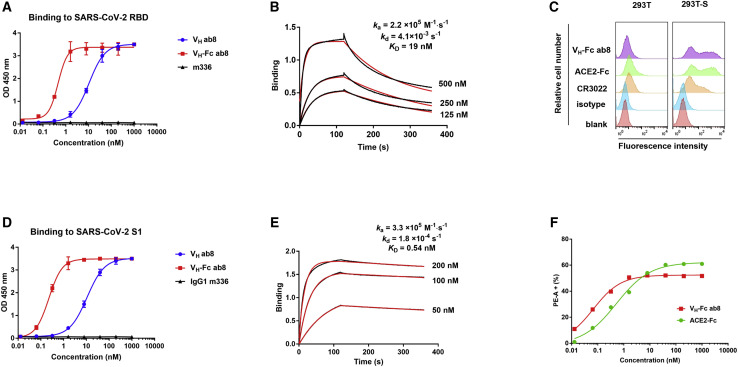
VH ab8 bound to SARS-CoV-2 RBD and S1 with half-maximal binding concentrations (EC50s) of 10 nM as measured by ELISA (Figures 1A and 1D) and an equilibrium dissociation constant (KD) of 19 nM as measured by the biolayer interferometry (Blitz system) (Figure 1B). The relatively fast dissociation rate constant (kd = 4.1 × 10−3 S−1) was significantly (23-fold) decreased by the conversion to a bivalent Fc fusion format (kd = 1.8 × 10−4 S−1) (Figure 1E) resulting in high avidity. VH-Fc ab8 bound to SARS-CoV-2 RBD and S1 subunit of S protein with EC50s of 0.40 nM and 0.20 nM, respectively, and a KD of 0.54 nM (Figure 1E). It specifically bound to 293T cells expressing S, but not to control 293T cells (Figures 1C and S2 A). The binding of VH-Fc ab8 was higher than that of IgG1 CR3022, an anti-SARS-CoV antibody cross-reactive with SARS-CoV-2 (Tian et al., 2020). The VH-Fc ab8’s half-maximal fluorescence-activated cell sorting (FACS) measured binding concentration (FC50) of 0.07 nM was higher than that of recombinant human ACE2-Fc (FC50 = 0.52 nM) (Figure 1F). These data demonstrate that ab8 selected by an isolated RBD can bind to cell surface associated native S trimer. The binding of VH-Fc ab8 to the S protein was significantly improved compared to that of the VH ab8 through avidity effect.
Figure 1.
Binding of VH ab8 and VH-Fc ab8 to Recombinant SARS-CoV-2 RBD and S1 Proteins and Cell Membrane-Associated S
(A and D) VH and VH-Fc ab8 binding to recombinant RBD and S1 proteins measured by ELISA. The MERS-CoV antibody IgG1 m336 was used as a negative control. Experiments were performed in duplicate and the error bars denote ± SD, n = 2. (D) shows concentration-dependent binding of VH-Fc ab8 and ACE2-Fc to 293T-S cells.
(B and E) Kinetics of VH ab8 (B) and VH-Fc ab8 (E) binding to RBD.
(C) Binding of VH-Fc ab8, ACE2-Fc, and IgG1 CR3022 to S transiently transfected 293T cells (293T-S). The 293T cells without transfection serve as a control. Antibodies or proteins were evaluated at a concentration of 1 μM.
See also Figure S2A.
Figure S2.
Concentration-Dependent Binding of VH-Fc ab8 and ACE2-Fc to Cell Surface-Associated SARS-CoV-2 S, Evaluation of Competition of ACE2 and VH ab8 by ELISA and FACS, and Test of the Conformation Integrity of RBD Mutants by Using a Polyclonal Antibody and Monoclonal Antibody CR3022, Related to Figures 1, 2, and 3
(A) Cells were incubated with serially diluted antibodies or ACE2-Fc and subsequently with PE conjugated anti-human Fc antibody for flow cytometry analysis. Percentage of PE-A+ cells were defined by the above gate strategy in FlowJ, representing the percentage of VH-Fc ab8 and ACE2-Fc bound 293T-S cells. (B) ACE2 blocking VH ab8 for binding to RBD by ELISA. RBD was coated to plate and 10 nM of VH ab8 in the presence of gradient concentration of ACE2 was added. Binding was detected by HRP conjugated anti FLAG tag antibody. (C) ACE2 blocking VH ab8 for binding to cell surface associated S. S transiently transfected 293T was incubated with 1 μM VH ab8 in the presence of various concentration of ACE2 (his tag). Binding of VH ab8 was detected by the PE conjugated anti FLAG tag antibody. (D) and (E) Binding of a mouse polyclonal anti-SARS-CoV-2 RBD antibody and IgG1 CR3022 to the RBD mutants. RBD mutants were coated to plate and two concentrations of polyclonal anti-RBD antibody and CR3022 were added. Binding was detected by HRP conjugated anti mouse (Fc) antibody and anti-human (Fc) antibody. Experiments were performed in duplicate and the error bars denote ± SD, n = 2.
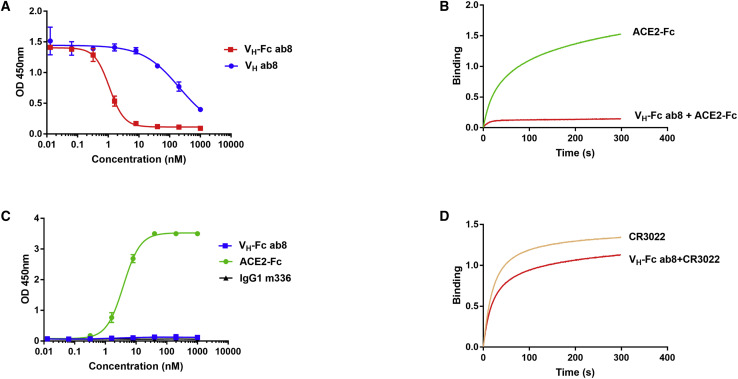
VH-Fc ab8 and VH ab8 Outcompete Human ACE2-Fc for Binding to RBD
Competition with human ACE2 for binding to RBD is a surrogate indicator for antibody neutralization activity. VH-Fc ab8 outcompeted human ACE2-Fc with a half-maximal inhibitory concentration (IC50) of 1.0 nM (Figure 2 A). Note that the VH-Fc ab8 was much more effective in outcompeting ACE2-Fc than VH ab8, consistent with its enhanced binding. ACE2 can also block VH ab8 for binding to RBD (Figure S2B) and cell surface associated S (Figure S2C). VH-Fc ab8 also significantly decreased the kinetics of ACE2 binding as measured by Blitz (Figure 2B). VH-Fc ab8 did not bind to the SARS-CoV RBD (Figure 2C) and did not compete with CR3022 for binding to RBD (Figure 2D). The CR3022 epitope is located in a conserved region on the RBD core domain distal from the ACE2 binding interface, as seen in the crystal structure of the Fab CR3022-RBD complex (Yuan et al., 2020). These results indicate that the ab8 epitope may overlap with the ACE2 binding site on RBD.
Figure 2.
Competition of VH-Fc ab8 and VH ab8 with ACE2, CR3022 for Binding to SARS-CoV-2 RBD, and Lack of Binding of VH-Fc ab8 to SARS-CoV S1
(A) Competition of VH-Fc ab8 and VH ab8 with ACE2 for binding to SARS-CoV-2 RBD. RBD was coated and incubated with 5-fold serially diluted VH-Fc ab8 and VH ab8 in the presence of 2 nM ACE2-mFc (mouse Fc).
(B) Inhibition of ACE2 binding to RBD by VH-Fc ab8 as measured by Blitz.
(C) Lack of binding to SARS-CoV S1 as tested by ELISA. SARS-CoV S1 was coated and incubated with VH-Fc ab8.
(D) Competition between VH-Fc ab8 and CR3022 measured by Blitz. ELISA Experiments were performed in duplicate and the error bars denote ±SD.
See also Figures S2B and S2C.
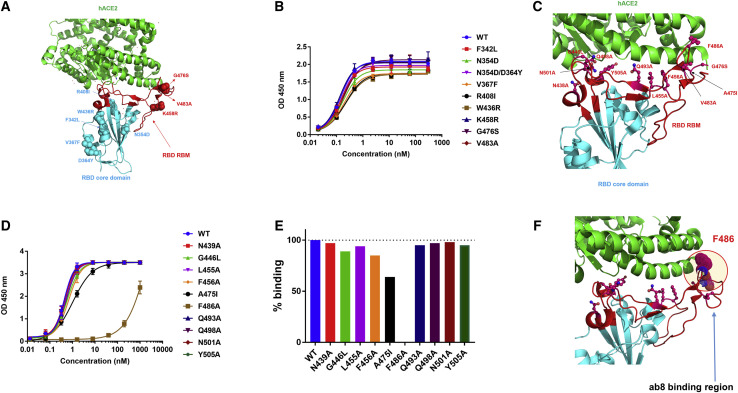
VH-Fc ab8 Binds to SARS-CoV-2 RBD Mutants Found in Patients; an Alanine Scanning Mutation in the Distal Loop Tip of the Receptor Binding Motif Decreases Its Binding
Currently, nine prevalent RBD mutants were found in COVID-19 patients (Priyanka et al., 2020). Six of these mutations (F342L, N354D. N354D/D364Y, V367F, R408I, and W436R) are located in the RBD core domain and three, K458R, G476S and V483A are in the receptor binding motif (RBM) (Figure 3 A). VH-Fc ab8 bound to all mutants similarly to wild-type RBD as measured by ELISA (Figure 3B). To map the ab8 epitope, we also generated several mutations in non-conserved positions compared to SARS-CoV spanning the footprint of ACE2 on RBM (N439A, G446L, L455A, F456A, A475I, F486A, Q493A, Q498A, N501A, and Y505A) (Figure 3C). Most of these mutants retained VH-Fc ab8 binding except F486A, F456A, and A475I (Figures 3D and 3E). The F486A significantly decreased binding without affecting the overall RBD conformation (Figures S2C and S2D) indicating that F486 directly interacts with ab8. The F456A and A475I mutations decreased the binding by 15% and 40%, respectively, but they also affected the RBD conformation (Figures S2C and S2D). These results suggest that a portion of the VH ab8 epitope could be in the RBM distal loop tip where the F486 is located (Figure 3F).
Figure 3.
Epitope Mapping for VH-Fc ab8 by Using Naturally Occurring RBD Mutants from Circulating SARS-CoV-2 Isolates and by Alanine Scanning
(A) Mapping of natural RBD mutants to RBD/ACE2 3D structure (PDB: 6M0J). RBD and ACE2 are represented as cyan and green cartoons with RBM highlighted by red color. The RBD mutants are represented by cyan (core domain mutants) and red (RBM) spheres.
(B) Binding of VH-Fc ab8 to those RBD mutant as measured by ELISA. Experiments were performed in duplicate, and the error bars denote ± SD, n = 2.
(C) Design of Ala scanning mutants to explore the ab8 epitope. RBD/ACE2 structure is based the same PDB as (A). Non-conservative residues spanning ACE2 footprint on RBD compared to SARS-CoV are selected and depicted by stick and sphere representations.
(D) VH-Fc ab8 binding to SARS-CoV-2 RBD alanine mutants as tested by ELISA. ELISA procedure is similar to the above described.
(E) Normalized signals of VH-Fc ab8 binding to those RBD mutants compared to the WT RBD at the concentration of 1.6 nM derived from (D).
(F) Representation of portions of ab8 binding region on RBD based on the epitope mapping ELISA results. F486 in the distal RBM loop is the plausible direct contact residue for ab8.
See also Figures S2D and S2E.
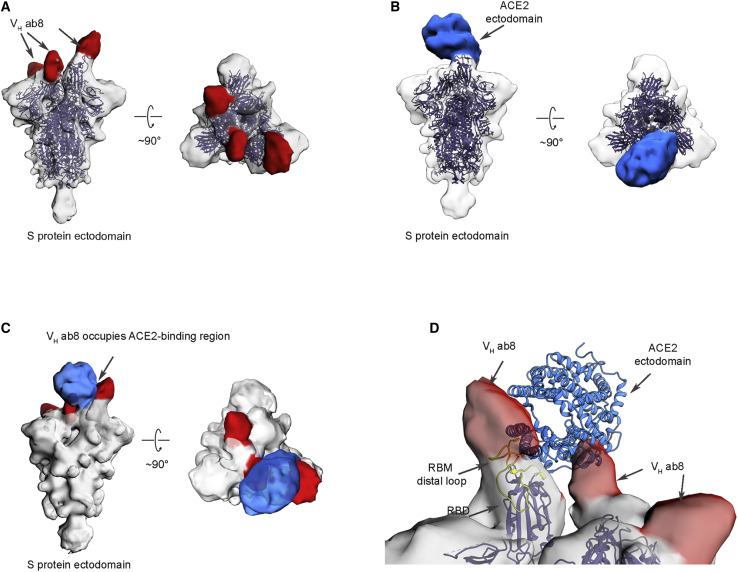
Electron Microscopic Analysis of the SARS-CoV-2 S Protein Ectodomain Bound to VH ab8
To explore structural aspects of SARS-CoV-2 neutralization by VH ab8, we performed negative stain electron microscopic analysis (Figure S3 ) of the complex formed between the S protein ectodomain and VH ab8 or soluble ACE2 (Figure 4 ). The density maps showed that both VH ab8 and ACE2 were in a quaternary conformation in which two of the protomers in the trimer are in the “down” conformation with the third one in the “up” conformation (Figures 4A and 4B), similar to the quaternary conformation of the reported ACE2-bound S ectodomain (PDB: 6VYB). One molecule of the VH ab8 was observed bound to each RBD domain (Figure 4A). In the ACE2-S complex, one molecule of ACE2 was bound to the S protein trimer, straddling one “up” and one “down” RBD region (Figure 4B). There appears to be a noticeable shift of the “up” RBD domain when it is bound to VH ab8 (Figure 4A). This shift is not observed when ACE2 is bound to the trimer (Figure 4B). Superposition of the two density maps reveals that the binding site of VH ab8 directly overlaps with the ACE2 one, precluding simultaneous occupancy on the S protein ectodomain (Figure 4C). We also found that when ACE2 was added subsequent to the addition of VH ab8, only the VH ab8 bound state was observed, further confirming the ACE2 competition with VH ab8. To better understand the spatial relationship between the site of VH ab8 binding and that of ACE2 binding, we created a molecular model for ACE2 bound S trimer by aligning the RBD region of the crystal structure of SARS-CoV-2 RBD bound ACE2 (PDB: 6M0J) (Lan et al., 2020) to the “up” RBD region in the cryo-EM structure of the trimer (PDB: 6YVB) (Wrapp et al., 2020). Superposition of this chimeric structure with the density map of VH ab8-bound S protein trimers reveals that the bound ACE2 has extensive overlap with the space occupied by bound VH ab8 (Figure 4D). The direct spatial overlap between bound VH ab8 and ACE2 provides a structural mechanism for the observed effect of ab8 on blocking ACE2 binding. The structural findings also showed that the RBM distal loop, which has F486 at its tip, is directly covered by the footprint of the bound VH ab8, consistent with the epitope mapping results showing that F486 is a direct contacting residue for ab8.
Figure S3.
Collection and Analysis of Electron Microscopic Data, Related to Figure 4
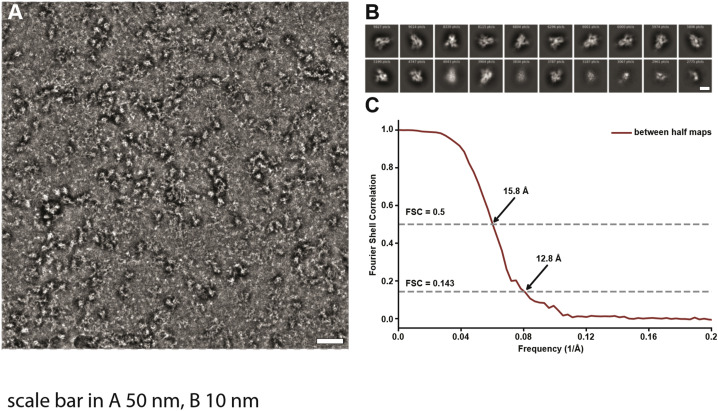
(A) Representative raw micrograph of the SARS-CoV-2 S protein ectodomain complex with VH ab8. Scale bar 50 nm. (B) Selected 2D class averages. Scale bar 10 nm. (C) Plot of Fourier Shell Correlation (FSC) between maps constructed from two randomly selected halves of the particle projection images.
Figure 4.
Electron Microscopic Analysis of the SARS-CoV-2 S Protein Ectodomain Complexed with VH ab8
(A) Side and top views of the density map of S protein ectodomain (shown in gray) in complex with VH ab8. The density that we associate with the bound VH domain is colored red. The open-state structure of the SARS-CoV-2 S protein ectodomain (PDB: 6VYB, blue color ribbon) fits well into the map with the exception of the tip of the RBD from the “up” protomer. There appears to be a slight outward shift in the VH ab8 complex.
(B) Side and top views of the density map of S protein ectodomain in complex with soluble human ACE2 domain, with density for bound ACE2 shown in blue.
(C) Superposition of the density maps from (A) and (B).
(D) A closer view of the binding site that incorporates the known atomic model for the structure of the ACE2 complex with the RBD in the “up” conformation, delineating the regions of contact with the VH density. A ribbon representation of the RBM distal loop and the F486 side chain are highlighted in yellow.
See also Figure S3.
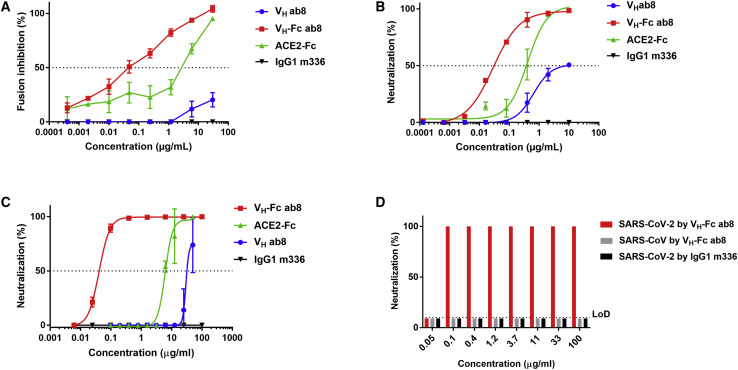
Potent Neutralization of SARS-CoV-2 by VH-Fc ab8 In Vitro
We used four different assays to evaluate VH-Fc ab8 mediated inhibition of SARS-CoV-2 infection in vitro: a β-galactosidase (β-Gal) reporter gene-based quantitative cell-cell fusion assay (Xiao et al., 2003); an HIV-1 backbone-based SARS-CoV-2 pseudovirus assay (Zhao et al., 2013); and two different replication-competent virus neutralization assays (a luciferase reporter gene assay and a microneutralization [MN]-based assay) (Scobey et al., 2013; Yount et al., 2003). VH-Fc ab8 inhibited cell-cell fusion much more potently than VH ab8 (Figure 5 A). The inhibitory activity of VH-Fc ab8 was also higher than that of ACE2-Fc. The control anti MERS-CoV antibody IgG1 m336 did not show any inhibitory activity. VH-Fc ab8 neutralized pseudotyped SARS-CoV-2 virus (IC50 = 0.03 μg/mL) more potently than ACE2-Fc (IC50 = 0.40 μg/mL) and VH ab8 (IC50 = 0.65 μg/mL) (Figure 5B). The pseudovirus neutralization IC50 for ACE2-Fc in our assay is comparable to the one reported by Lei et al. (2020) (0.03–0.1 μg/mL). Interestingly, the maximum neutralization by VH ab8 was only 50% compared to the 100% by VH-Fc ab8 and ACE2-Fc, which was also observed for another antibody S309 (Pinto et al., 2020). The complete neutralization by VH-Fc ab8/ACE2-Fc emphasizes the role of bivalency and related avidity in neutralization (Klasse and Sattentau, 2002). Furthermore, in the reporter gene assay VH-Fc ab8 neutralized live SARS-CoV-2 with an IC50 of 0.04 μg/mL (Figure 5C), which is much lower than that for ACE2-Fc (IC50 of 6.1 μg/mL) and VH ab8 (IC50 = 29 μg/mL). ACE2-Fc seemed to be much less potent against the live virus compared to the pseudovirus, which is also observed by others (IC50 = 12.6 μg/mL) (Case et al., 2020) and may relate to the S expression levels and RBD/S conformation on the virus surface. We also confirmed the high VH-Fc ab8 live virus neutralization potency by a microneutralization (MN) assay-100% neutralization (NT100) at 0.1 μg/mL (Figure 5D). The NT100 from the MN assay (0.1 μg/mL) was close to the IC100 (0.2 μg/mL) from the reporter gene assay suggesting consistency in the live virus neutralizing activity of VH-Fc ab8 obtained with two independent assays at two different laboratories. These results suggest that VH-Fc ab8 is a potent neutralizer of SARS-CoV-2, which correlates with its strong competition with ACE2 for binding to RBD.
Figure 5.
Inhibition of Cell-Cell Fusion and Neutralization of Pseudotyped and Authentic SARS-CoV-2 by VH-Fc ab8 and VH ab8
(A) Inhibition of cell fusion between 293T-S and 293T-ACE2 cells by VH ab8, VH-Fc ab8, and ACE2-Fc.
(B) Neutralization of SARS-CoV-2 pseudovirus by VH ab8, VH-Fc ab8, and ACE2-Fc.
(C) Neutralization of live SARS-CoV-2 tested in the nLuc reporter assay.
(D) Neutralization of live virus by a microneutralization assay. Experiments were performed in duplicate and the error bars denote ±SD, n = 2.
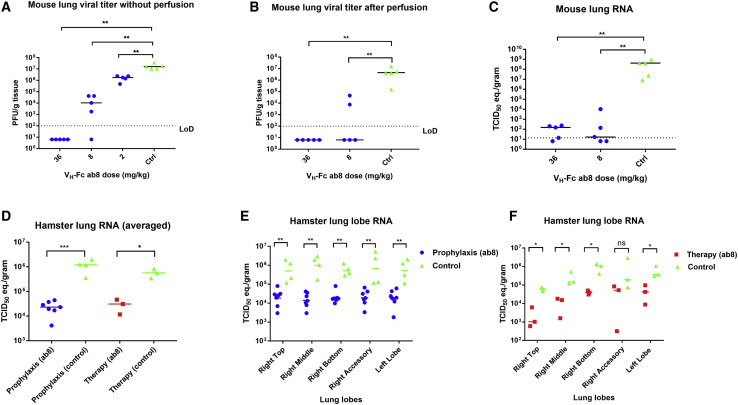
High Prophylactic Efficacy of VH-Fc ab8 in a Mouse ACE2-Adapted SARS-CoV-2 Infection Model
To evaluate the prophylactic efficacy of VH-Fc ab8 in vivo, we used a recently developed mouse ACE2-adapted SARS-CoV-2 infection model, in which wild-type BALB/c mice are challenged with SARS-CoV-2 carrying two mutations Q498T/P499Y at the ACE2 binding interface in the RBD (Dinnon et al., 2020). It was shown that in this model, the aged BALB/c mice exhibited more clinically relevant phenotypes than those seen in hACE2 transgenic mice (Dinnon et al., 2020). Groups of 5 mice each were administered 36, 8, and 2 mg/kg VH-Fc ab8 prior to high titer (105 plaque-forming unit [pfu]) SARS-CoV-2 challenge followed by measurement of virus titer in lung tissue 2 days post infection (dpi). VH-Fc ab8 effectively inhibited SARS-CoV-2 in the mouse lung tissue in a dose-dependent manner (Figure 6 A). There was complete neutralization of infectious virus at the highest dose of 36 mg/kg, and statistically significant reduction by 1,000-fold at 8 mg/kg. Remarkably, even at the lowest dose of 2 mg/kg it significantly decreased virus titer by 10-fold (two-tailed, unpaired t test, p = 0.0075). To exclude possible effects of residual ab8 on viral titration, we performed another experiment in which mouse lungs were perfused with 10 mL of PBS before harvesting for titration. The perfusion did not affect to any significant degree the infectious virus in the lungs (Figure 6B). The VH-Fc ab8 completely neutralized the virus in the lungs at 36 mg/kg and significantly reduced infectious virus at 8 mg/kg. VH-Fc ab8 also reduced viral RNA in the lungs (Figure 6C). These results demonstrate the neutralization potency of VH-Fc ab8 in vivo. They also suggest that the double mutations Q498T/P499Y on RBD did not influence VH-Fc ab8 binding and contribute to the validation of the mouse-adapted SARS-CoV-2 model for evaluation of neutralizing antibody efficacy.
Figure 6.
Evaluation of the Prophylactic Efficacy of VH-Fc ab8 in a Mouse ACE2-Adapted Model and Both Prophylactic and Therapeutic Efficacy in a Hamster Model of SARS-CoV-2 Infection
(A) VH-Fc ab8 inhibited mouse ACE2-adapted SARS-CoV-2 in wild-type BALB/c mice (two-tailed, unpaired t test, ∗∗p < 0.01).
(B) The same experiments as (A) except that the mice lung was perfused before viral titration (Mann-Whitney U test, ∗∗p < 0.01).
(C) The viral RNA level change in the lung in the same mice of (B) as quantified by RT-qPCR and presented as TCID50 equivalents (Mann-Whitney U test, ∗∗p < 0.01).
(D–F) Evaluation of the prophylactic and therapeutic efficacy of VH-Fc ab8 in the hamster model. Hamsters were injected intraperitoneally with 10 mg/kg of VH-Fc ab8 antibody either 1 day before (prophylaxis) or 6 h after (therapy) intranasal challenge of 1 × 105 TCID50 of SARS-CoV-2. (D) The decrease of viral RNA in the hamster lung after averaging all lung lobes. (E and F) The decrease of viral RNA in hamster lung lobes: prophylaxis and therapy, respectively (Mann-Whitney U test, not significant [ns]: p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
See also Figures S4 and S5.
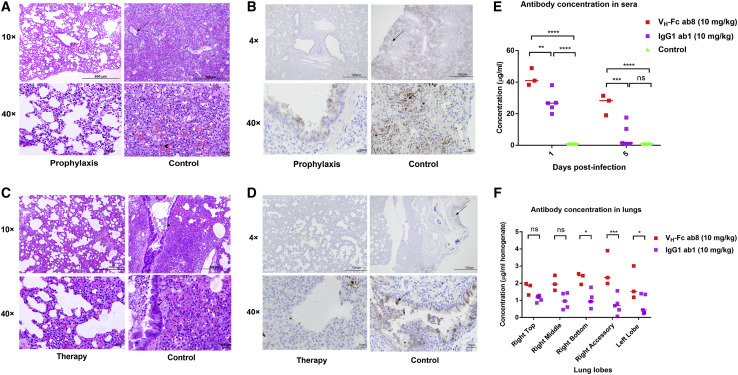
VH-Fc ab8 Exhibited Both Prophylactic and Therapeutic Efficacy in a Hamster Model of SARS-CoV-2 Infection
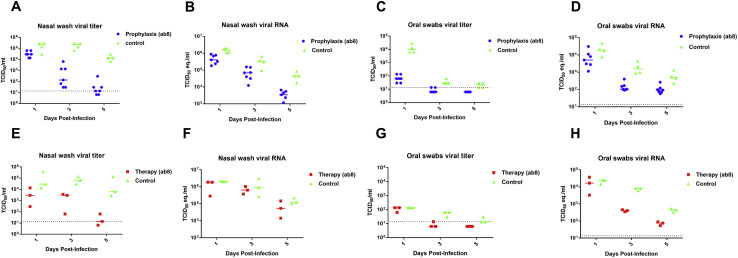
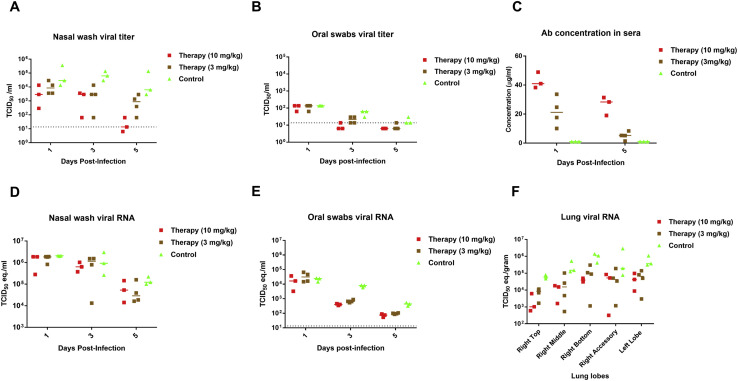
Recently, hamsters were demonstrated to recapitulate clinical features of SARS-CoV-2 infection (Chan et al., 2020; Imai et al., 2020). To evaluate the VH-Fc ab8 efficacy in hamsters, it was intraperitoneally administered either 24 h before (prophylaxis) or 6 h after (therapy) intranasal 105 TCID50 virus challenge. In the therapeutic group, the rationale for administration of the antibody 6 h post viral infection is based on the replication cycle length of 5–6 h after initial infection for SARS-CoV in VeroE6 cells (Keyaerts et al., 2005). Six h after challenge with a high dose of 105 fifty-percent tissue culture infective dose (TCID50), approximately the same number of susceptible cells could become infected and likely produce much more infectious virus, which would need to be neutralized by the antibody to prevent subsequent cycles of infection. Nasal washes and oral swab at 1, 3, and 5 dpi and different lung lobes at 5 dpi were collected. VH-Fc ab8 decreased viral RNA by 1.7 log in the lung when administered prophylactically. The lung viral RNA decrease in the therapeutic groups was slightly lower (by 1.2 log) (Figure 6D). Interestingly, the viral RNA load in the therapeutic groups was to some extent tissue location-dependent (Figure 6F). The variation of the viral load in different lung lobes may relate to nonuniform antibody transport and viral spread inside the lung. Remarkably, VH-Fc ab8 alleviated hamster pneumonia and reduced the viral antigen in the lung (Figures 7A and 7C, H&E staining, and 7B and 7D, immunohistochemistry). The control hamsters exhibited severe interstitial pneumonia characterized by extensive inflammatory cell infiltration, presence of type II pneumocytes, alveolar septal thickening, and alveolar hemorrhage. Both prophylactic and therapeutic treatment of VH-Fc ab8 reduced the lesions of alveolar epithelial cells, focal hemorrhage, and inflammatory cells infiltration. VH-Fc ab8 also reduced the shedding from mucosal membranes including in nasal washes and oral swabs (Figure S4 ). The decrease in viral RNA in nasal washes and oral swabs were not as large as the decrease observed in the lung tissue, similar to a recent finding in hamsters (Imai et al., 2020). Overall, the prophylactic treatment was more effective than the therapeutic treatment in decreasing viral load in nasal washes and oral swabs. Notably, prophylactic administration of VH-Fc ab8 effectively reduced the infectious virus in the oral swab at 1 dpi, while the post-exposure treatment did not (Figures S4C and S4G). Interestingly, viral reduction (except the viral titer in the oral swab at 1 dpi) was more effective at 3 and 5 dpi compared to that at 1 dpi, likely due to the infection peak occurring before day 3 as reported in hamsters (Sia et al., 2020). A striking finding is that VH-Fc ab8 given therapeutically at as low dose as 3 mg/kg can still decrease viral loads in the lung, nasal washes, and oral swabs (Figure S5 ).
Figure 7.
Histopathology of Hamster Lung Stained by H&E and Immunohistochemistry (IHC); Comparison of Antibody Concentrations in the Hamster Lung and Sera between VH-Fc ab8 and IgG1 ab1
(A and C) Reduced pathological changes in lung tissue lobe with VH-Fc ab8 treatment. H&E staining of treated and control lung lobes in hamsters challenged with SARS-CoV-2. Arrows showed inflammatory cells and arrow head for alveolar hemorrhages.
(B and D) Prophylaxis and post-infection treatment with VH-Fc ab8 decreased SARS-CoV-2 antigen staining in lung lobes of hamsters. IHC detection of the nucleocapsid antigen of VH-Fc ab8 prophylactically treated (B) and post-exposure treatment (D) and control hamster lungs following SARS-CoV-2 challenge. Arrow indicates nucleocapsid positive cells (brown) in lungs lobes of hamsters at day 5 post-infection.
(E and F) Comparison of VH-Fc ab8 and IgG1 ab1 concentration in the lung and sera of hamsters receiving post-exposure treatment of a dose of 10 mg/kg (two-way ANOVA analysis followed by Tukey test, ns, p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Figure S4.
Detection of Infectious Virus and Viral RNA in Hamster Nasal Washes and Oral Swabs, Related to Figure 6
Hamsters were injected intraperitoneally with 10 mg/kg of VH-Fc ab8 antibody either one day before (prophylaxis) or six hours after (therapy) intranasal challenge of 1 × 105 TCID50 of SARS-CoV-2. Untreated hamsters were kept as a control. Nasal washes and oral swabs were collected at day one, three and five post infection (dpi) for virus titer titration by viral TCID50 assays and viral RNA quantification by RT-qPCR. (A) and E). Nasal washes viral titer in un-treated (control), pre-infection (prophylaxis) treatment and post-infection (therapy) treatment hamsters. (B) and F). Nasal washes viral RNA levels in un-treated, pre-treated and post- treated hamsters. (C) and (G) Oral swab viral titer in un-treated, pre-treated and post-treated hamsters. Note that the prophylactic treatment of VH-Fc ab8 largely decreased the viral tier in the oral swabs at one dpi, while there is almost no effect for the post-infection treatment. (D) and (H) Oral swab viral RNA levels in un-treated, pre-treated and post-treated hamsters.
Figure S5.
Post-exposure Treatment Efficacy of VH-Fc ab8 at Two Different Doses in the Hamster Model, Related to Figure 6
VH-Fc ab8 at doses of 10 mg/kg or 3 mg/kg was administered i.p. 6 h after virus intranasal challenge. The hamster shedding including nasal washes and oral swabs were collected at 1, 3, 5 dpi. All hamsters were euthanized on 5 dpi. At the euthanasia, lungs (different lobes) were collected viral RNA quantification by RT-qPCR. (A) and (D) Nasal washes viral titer and viral RNA in un-treated (control), 3 mg/kg and 10 mg/kg post-infection treated hamsters. (B) and (E) Oral swab viral titer and viral RNA in un-treated (control), 3 mg/kg and 10 mg/kg post-infection treated hamsters. (C) Comparison of antibody concentrations in hamster sera for those two doses. Hamsters were bled at one and five dpi for measuring antibody concentrations in sera by SARS-CoV-2 S1 ELISA. Sera was diluted 1:100 and binding was detected by using the goat anti human IgG-HRP. (F). Viral RNA levels in different lung lobes. RNA quantity was presented as the TCID50 equivalence.
We measured the VH-Fc ab8 concentrations at both doses (10 and 3 mg/kg) in the sera at 1 dpi and 5 dpi in the post-exposure treatment groups (Figure S5C). The higher dose (10 mg/kg) resulted in higher antibody concentration and better inhibitory activity than the lower dose (3 mg/kg). The relatively high concentration of VH-Fc ab8 5 days after administration also indicates good pharmacokinetics. Furthermore, we also compared the VH-Fc ab8 concentration in both the sera and lung with that of IgG1 ab1, which has a similar affinity to SARS-CoV-2 and similar degree of competition with the receptor ACE2 as VH-Fc ab8 (Li et al., 2020a). We found that the concentration of VH-Fc ab8 in hamster sera is significantly higher than that of IgG1 ab1 at 1 and 5 dpi after post-exposure administration of the same dose of 10 mg/kg (Figure 7E), possibly indicating more effective delivery of VH-Fc ab8 from the peritoneal cavity to the blood than that of IgG1 ab1. We also found that the VH-Fc ab8 concentration in all hamster lung lobes was higher than that of the IgG1 ab1 (Figure 7F), suggesting that VH-Fc ab8 appears to penetrate the lung tissue more effectively than IgG1 ab1. These results indicate that the in vivo delivery of VH-Fc ab8 may be more effective than that of full-size antibodies in an IgG1 format.
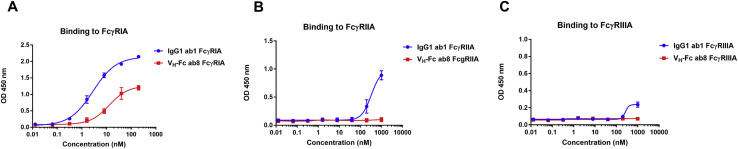
VH-Fc ab8 Does Not Aggregate and Does Not Bind to 5,300 Human Membrane Proteins
The VH-Fc ab8 propensity for aggregation was measured at 37°C by dynamic light scattering (DLS), which detects particle size distributions in the nanometer range (Stetefeld et al., 2016). It displayed a single peak at 11.5 nm which is the size of a monomeric VH-Fc protein (Figure S6 A). The absence of large-size peaks corresponding to large molecular weight species (aggregates) in solution indicates that VH-Fc ab8 is highly resistant to aggregation at high concentration (4 mg/mL) and relatively long times of incubation (6 days) at 37°C. The VH-Fc ab8 propensity for aggregation was also evaluated by size exclusion chromatography (SEC), which showed that >96% of VH-Fc ab8 was eluted in a peak at a position corresponding to a monomeric state with a molecular weight of 80 kDa (Figure S6B).
Figure S6.
Absent or Very Low Aggregation and High Specificity of Binding of VH-Fc ab8, Related to STAR Methods
(A) Evaluation of the aggregation of VH-Fc ab8 by DLS. VH-Fc ab8 (4 mg/ml) buffered in PBS was incubated at 37°C. On day 0, day 1 and day 6, samples were taken out for DLS measurement on Zetasizer Nano ZS ZEN3600 (Malvern Instruments Limited, Westborough, MA) to determine the size distribution. All measurements were repeated by three times. (B) Evaluation of VH-Fc ab8 aggregation by SEC. Size exclusion was performed by loading 0.22 μm membrane-filtered proteins (150 ul, 1.5 mg/mL) onto the Superdex 200 increase 10/300 GL column. Protein was eluted by PBS buffer in a flow rate of 1.5 mL/min. The arrows indicate the peaks of the MW standards in PBS. (C) Lack of non-specific binding measured by a Membrane Proteome Array (MPA). Specificity testing of VH-Fc ab8 (20 μg/ml) was performed using the MPA platform which comprises 5,300 different human membrane proteins, each overexpressed in live cells. To ensure data validity, each array plate contained positive (SARS-CoV-2 S) and negative (empty vector) controls.
Antibody nonspecificity and polyreactivity can be an obstacle for developing an antibody into a clinically useful therapeutic. Polyreactivity may not only cause off-target toxicities and interfere with normal cellular functions, but may also reduce antibody half-life (Chuang et al., 2015). To test for potential polyreactivity of VH-Fc ab8, a membrane proteome array (MPA) platform was used, in which 5,300 different human membrane protein clones were separately overexpressed in 293T cells in a matrix array achieving a high-throughput detection of binding by FACS. VH-Fc ab8 did not bind to any of those proteins (Figure S6C), demonstrating its lack of polyreactivity and nonspecificity. Interestingly, we did not detect VH-Fc ab8 binding to the human FcγRIA, which is probably due to the relatively low expression level of FcγRIA on HEK293T cell surface without concomitant expression of the common γ chain (van Vugt et al., 1996). In addition, we found that VH-Fc ab8 bound to the FcγRs much weaker than IgG1 (Figure S7 ), likely due to the different conformation in the lower hinge region for Fc fusion proteins compared to that of IgG1s (Ying et al., 2014b). For the Fc fusion proteins (even with the same hinge sequence as IgG1), binding to FcγRs may be different from that of IgG1 and can be affected by the fusion partners (Lagassé et al., 2019). The importance of antibody binding to FcγRs for therapeutic or prophylactic efficacy or toxicity in SARS-CoV-2 infection is unknown.
Figure S7.
Binding of VH-Fc ab8 to Human FcγRs Measured by ELISA, Related to STAR Methods
Recombinant FcγRs ectodomains (100 ng) were coated, and biotinylated VH-Fc ab8 or IgG1 ab1 was added. Binding was detected by Streptavidin HRP. Experiments were performed in duplicate and the error bars denote ± SD, n = 2.
Discussion
Neutralizing mAbs are promising for prophylaxis and therapy of SARS-CoV-2 infections. Recently, many potent neutralizing antibodies from COVID19 patients were identified that neutralize pseudovirus with IC50s ranging from 1 to 300 ng/mL, and replication-competent SARS-CoV-2 with IC50s from 15 to 500 ng/mL (Cao et al., 2020; Ju et al., 2020; Rogers et al., 2020; Shi et al., 2020; Zost et al., 2020). By comparison, the VH-Fc ab8 reported here exhibited comparable or better neutralizing potency against SARS-CoV-2 pseudovirus and live virus (IC50s of 30 ng/mL and 40 ng/mL, respectively). Of note, IC50s can vary widely between different assays and laboratories because there is no generally accepted standardized assay. In addition, there are many factors that contribute to potency and efficacy in vivo. Animal models are a more comprehensive and likely more reliable predictor of potential efficacy in humans than in vitro neutralization assays.
To our knowledge, VH-Fc ab8 is the first human antibody domain whose activity was validated in two animal models. In the mouse ACE2-adapted SARS-CoV-2 infection model, VH-Fc ab8 significantly decreased infectious virus by 10-fold at 2 dpi even at a very low dose of 2 mg/kg (Figure 6A). It also exhibited both prophylactic and therapeutic efficacy in a hamster model. It not only reduced the viral load in the lung and alleviated pneumonia, but it also reduced shedding in the upper airway (nasal washes and oral swab), which could potentially reduce transmission of SARS-CoV-2. Impressively, VH-Fc ab8 was active therapeutically, even at 3 mg/kg. The finding that VH-Fc ab8 persisted for 4 days post administration at significant levels indicates that the pharmacokinetics of VH-Fc ab8 is comparable to that of a full size antibody; the half-lives of Fc fusion proteins were reported to vary from those of IgG1s and can range from hours to days (Unverdorben et al., 2016). The molecular weight of VH-Fc ab8 (80 kDa) is half of that of full-size IgG1 that suggests an advantage in terms of smaller quantities needed to be produced compared to those for IgG1s to reach similar number of molecules and efficacy. In addition, it was shown that decreasing binder’s size exponentially increases its diffusion through normal and tumor tissues (Jain, 1990). Thus, decreasing the size 2-fold can increase diffusion through tissues by 4-fold. We found that after administration at the same dose, the concentration of VH-Fc ab8 was higher than that of IgG1 ab1 in both hamster sera and lung tissue. This result might suggest that the VH-Fc ab8 diffusion from the peritoneal cavity to the blood and penetration of lung may be faster than that of IgG1 ab1. This may further explain its efficacy at low doses in animals. Although the low dose showed efficacy in the small animal models, it should be noted that in humans higher doses could be required to achieve comparable degree of efficacy. Another caveat is that in the hamster post-exposure experiment, the VH-Fc ab8 was administered at a time (6 h) when the first round of virus replication was likely completed (Keyaerts et al., 2005), but before the infection peak at 1–2 days (Sia et al., 2020). Because it inhibits infection of new cells, its administration at around the infection peak or after may not be as effective unless it also kills infected cells in vivo, which is under investigation.
Recently, antibody domains including human VH and camelid VHH were reported having varying neutralization potency (Chi et al., 2020; Sun et al., 2020; Wrapp et al., 2020; Wu et al., 2020a). Compared to those domains, VH-Fc ab8 is unique in terms of potency, aggregation resistance, and specificity. VH-Fc ab8 exhibited good developability properties including stability at high concentrations and long incubation at 37°C, as well as absence or very low aggregation. In addition, VH-Fc ab8 did not bind to the human cell line 293T even at high concentration (1 μM) which is ~1,754-fold higher than its Kd indicating absence of non-specific binding to many membrane-associated human proteins. A similar result was obtained by the membrane protein array assay showing that VH-Fc ab8 did not bind to any of 5,300 human membrane-associated proteins, indicating its lack of non-specificity and thus low potential for off-target toxicity when used in vivo. Unlike camel VHHs, the VH ab8 sequence is fully human and therefore likely less immunogenic than that of camelid VHHs.
Multiple structures are now available for the SARS-CoV-2 S protein trimer in complex with various neutralizing antibodies, offering insight into antigenic epitopes and inhibitory mechanisms critical for S protein neutralization. Epitopes on the SARS-CoV-2 S protein RBD have emerged as effective targets, as evidenced by the action of several RBD binding antibodies including CR3022, B38, C105, CB6, H014, and S309 (Barnes et al., 2020; Lv et al., 2020; Pinto et al., 2020; Shi et al., 2020; Wu et al., 2020b). While B38, C105, and CB6 directly compete with ACE2 for binding sites on the RBD surface, H014 occupies a position distinct from these binding sites, precluding ACE2 binding via steric inhibition (Lv et al., 2020). S309 targets the RBD of the S protein both in closed and open S protein conformations, exhibiting a different mechanism of neutralization (Pinto et al., 2020). A recent study of the structure of the S protein trimer in complex with the nanobody H11-D4 (PDB: 6Z43) revealed full occupancy of the nanobody on all three RBDs in a “one up and two down” conformation (Huo et al., 2020), similar to what we report here. Our structural analysis demonstrates that the location of the VH ab8 bound to the trimeric S ectodomain directly overlaps the region that would be occupied by ACE2 when bound to the S protein. The ACE2 blocking is likely the major mechanism of the VH-Fc ab8 neutralizing activity, which is significantly augmented by avidity effects due to its bivalency. The narrow neutralization concentration range in the live virus neutralization (10–200 ng/mL for 0%–100% neutralization) (Figure 5D) indicates a plausible cooperative neutralization mechanism, probably due to the synergistic binding of VH molecules in VH-Fc ab8 to RBDs. Due to its small size, VH may facilitate targeting occluded epitopes on RBD that are otherwise inaccessible to full-length IgGs, which is important because the SARS-CoV-2 S protein is conformationally heterogeneous, exposing neutralizing epitopes to varying degrees (Yan et al., 2020). The structural analysis shows that VH ab8 is able to simultaneously target all three RBD epitopes in both “up” and “down” conformations, which may provide a structural basis for a unique cooperative neutralization mechanism for VH-Fc ab8. VH-Fc ab8 with a long flexible linker between VH and Fc may allow two VH molecules to bind simultaneously two protomers in the same S trimer or cross-link two different protomers from different S trimers.
The ab8 epitope is distal to the CR3022 epitope, explaining its lack of competition with CR3022. The ab8 contact residue F486 (L472 in SARS-CoV) is not conserved that likely explains its lack of cross-reactivity to SARS-CoV. From the GISAID and NCBI databases, we found nine mutations in RBD with relatively high frequencies in current circulating SARS-COV-2. Six of them are in the core domain (F342L, N354D, N354D/D364Y, V367F, R408I, and W436R) and three in the RBM (K458R, G476S, and V483A). The core domain mutations are far away from the ab8 epitope, thus these mutations do not affect VH-Fc ab8 binding to RBD. Those three RBM mutations also did not affect ab8 binding although they are close to the ab8 epitope, suggesting these mutations may not affect ab8 neutralizing activity although neutralization of whole virus carrying these mutations is needed to definitely demonstrate this possibility. Interestingly, VH-Fc ab8 effectively inhibited the mouse ACE2-adapted SARS-CoV-2 with a Q498T/P499Y mutation in RBD, indicating that this double mutation also does not affect VH-Fc ab8 binding to RBD. These results suggest that VH-Fc ab8 may be a broadly cross-reactive SARS-CoV-2 neutralizing antibody.
In conclusion, we identified a fully human antibody VH domain that shows strong competition with ACE2 for binding to RBD and potent neutralization of SARS-CoV-2 in vitro and in two animal models. This potent neutralizing activity combined with its specificity and good developability properties warrants its further evaluation for prophylaxis and therapy of SARS-CoV-2 infection. Our elucidation of its unique epitope and mechanism of neutralization could also help in the discovery of more potent inhibitors and vaccines.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Phage display antibody library | ||
| VH phage library | This paper | N/A |
| Antibodies | ||
| VH ab8 | This paper | N/A |
| VH-Fc ab8 | This paper | N/A |
| IgG1 ab1 | This paper | N/A |
| IgG1 m336 | Ying et al., 2014a | N/A |
| IgG1 CR3022 | Yuan et al., 2020 | N/A |
| Streptavidin HRP | ThermoFisher | Cat# N100 |
| Anti FLAG HRP | Sigma-Aldrich | Cat# A8592-1MG; RRID: AB_439702 |
| Anti human Fc HRP | Sigma-Aldrich | Cat# A0170-1ML; RRID: AB_257868 |
| Anti mouse Fc HRP | Sigma-Aldrich | Cat# A0168-1ML; RRID: AB_257867 |
| Anti FLAG PE conjugate | Miltenyibiotec | Cat# 130-101-576; RRID: AB_2751038 |
| Anti human Fc PE conjugate | ThermoFisher | Cat# 12-4998-82; RRID: AB_465926 |
| Mouse pAb antiSARS2 RBD | Sino Biological | Cat# 40592-MP01 |
| Rabbit pAb antiSARS2 N | VIDO-Intevac | N/A |
| Goat anti Rabbit Ig HRP | Agilent | Cat# P044801-2; RRID: AB_2617138 |
| Alexa Fluor 488 anti human Fc | Jackson ImmunoResearch Labs | Cat# 109-545-098; RRID: AB_2337840 |
| Recombinant Proteins | ||
| SARS2 RBD-avi-his | This paper | N/A |
| SARS2 RBD-Fc | This paper | N/A |
| SARS2 S ectodomain | This paper | N/A |
| hACE2-hFc (human Fc) | This paper | N/A |
| hACE2-mFc (mouse Fc) | Sino Biological | Cat# 10108-H05H |
| hACE2-his | Sino Biological | Cat# 10108-H08H |
| SARS2 S1 | Sino Biological | Cat# 40591-V08B1 |
| SARS1 S1 | Sino Biological | Cat# 40150-V08B1 |
| SARS2 RBD N354D | Acrobiosystems | Cat# SPD-S52H5 |
| SARS2 RBDN354D/D364Y | Acrobiosystems | Cat# SPD-S52H3 |
| SARS2 RBD V367F | Acrobiosystems | Cat# SPD-S52H4 |
| SARS2 RBD R408I | Acrobiosystems | Cat# SPD-S52H8 |
| SARS2 RBD W436R | Acrobiosystems | Cat# SPD-S52H7 |
| SARS2 RBD F342L | Sino Biological | Cat# 40592-V08H6 |
| SARS2 RBD K458R | Sino Biological | Cat# 40592-V08H7 |
| SARS2 RBD N439A | This paper | N/A |
| SARS2 RBD G446L | This paper | N/A |
| SARS2 RBD L455A | This paper | N/A |
| SARS2 RBD F456A | This paper | N/A |
| SARS2 RBD A475I | This paper | N/A |
| SARS2 RBD G476S | This paper | N/A |
| SARS2 RBD V483A | This paper | N/A |
| SARS2 RBD F486A | This paper | N/A |
| SARS2 RBD Q493A | This paper | N/A |
| SARS2 RBD Q498A | This paper | N/A |
| SARS2 RBD N501A | This paper | N/A |
| SARS2 RBD Y505A | This paper | N/A |
| Recombinant FcγRIA | R&D systems | Cat# 1257-FC-050 |
| Recombinant FcγRIIA | R&D systems | Cat# 1330-CD-050 |
| Recombinant FcγRIIIA | R&D systems | Cat# 4325-FC-050 |
| Critical Commercial Reagents | ||
| Blitz Protein A sensor | ForteBio | Cat# 18-5010 |
| Blitz Streptavidin sensor | ForteBio | Cat# 18–5019 |
| QuikChange II XL Kit | Agilent | Cat# 200521 |
| β-galactosidase assay kit | G-Biosciences | Cat# 786-651 |
| Bright-Glo kit | Promega | Cat# E2610 |
| Nano-Glo Assay System | Promega | Cat# N1110 |
| QiaAmp Viral RNA mini kit | QIAGEN | Cat# 52904 |
| QuantiFast Probe RT-PCR kit | QIAGEN | Cat# 204454 |
| RNeasy kit | QIAGEN | Cat# 74104 |
| Deposited Data | ||
| Antibody nucleotide sequence | This paper | GenBank accession number: MT943599 |
| Bacterial and Virus Strains | ||
| TG1 | Lucigen | Cat#60502-1 |
| DH5α | Lucigen | Cat#60602-1 |
| vaccinia virus VTF7.3 | NIH | Cat#356 |
| vaccinia virus VCB21R | NIH | Cat# 3365 |
| SARS-CoV-2 (US_WA-1/2020) | CDC | N/A |
| SARS-CoV (Urbani) | CDC | N/A |
| Mouse ACE2 adapted SARS-CoV-2 | Dinnon et al., 2020 | N/A |
| SARS-CoV-UrbaninLuc | Hou et al., 2020 | N/A |
| SARS-CoV-2-SeattlenLuc | Hou et al., 2020 | N/A |
| SARSCoV2/Canada/ON/VIDO-01/2020 | VIDO-InterVac | N/A |
| Experimental Models: Cell lines | ||
| 293T | ATCC | ATCC® CRL-3216 |
| 293T-S | This paper | N/A |
| 293T-ACE2 | This paper | N/A |
| Expi293F | ThermoFisher | A14527 |
| HEK293F | ThermoFisher | R79007 |
| Vero-E6 | ATCC | ATCC® CRL-1586 |
| Experimental Models: Animals | ||
| BALB/c mice | Envigo | Cat#047 |
| Hamsters | Charles River Laboratory | https://www.criver.com/products-services/find-model/lvg-golden-syrian-hamster?region=24 |
| Oligonucleotides | ||
| 246 ESarbecoF1: ACAGGTACGTTAATAGTTAATAGCGT | Corman et al., 2020 | N/A |
| 247 ESarbecoR2: ATATTGCAGCAGTACGCACACA | Corman et al., 2020 | N/A |
| RT-PCR probe: 248E Sarbeco P1: ACACTAGCCATCCTTACTGCGCTTCG | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pNL4-3.luc.RE | NIH | Cat# 3418 |
| Plasmid: pcDNA3.1-RBD-his | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-Fc | This paper | N/A |
| Plasmid: pcDNA3.1-full length S | This paper | N/A |
| Plasmid: pcDNA3.1-full length human ACE2 | This paper | N/A |
| Plasmid: pcDNA3.1-ACE2-hFc | This paper | N/A |
| Plasmid: pcDNA3.1-ACE2-mFc | This paper | N/A |
| Plasmid: pSecTagB-VH-Fc ab8 | This paper | N/A |
| Plasmid: pComb3X-VH ab8 | This paper | N/A |
| Plasmid: pDR12-IgG1 ab1 | This paper | N/A |
| Plasmid: pCAGGS-S ecto 2P | BEI resources | NR-52394 |
| Plasmid: pcDNA3.1-RBD-mutant N439A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant G446L | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant L455A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant F456A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant A475I | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant G476S | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant V483A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant F486A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant Q493A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant Q498A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant N501A | This paper | N/A |
| Plasmid: pcDNA3.1-RBD-mutant Y503A | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism | GraphPad 7.0 | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator CC 2018 | Adobe | https://www.adobe.com |
| Snapgene | GSL Biotech LLC | https://www.snapgene.com/ |
| PyMoL | Schrödinger | https://pymol.org/2/ |
| FlowJ | FlowJo,V10, LLC | https://www.flowjo.com/solutions/flowjo/downloads |
| EPU automated acquisition | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/epu-software.html |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| RELION (3.1) | Scheres, 2012 | https://github.com/3dem/relion/releases/tag/3.1.0 |
| crYOLO (1.7.4) | Wagner et al., 2019 | https://pypi.org/project/cryolo/ |
| cryoSPARC live (v2.15.1) | Punjani et al., 2017 | https://cryosparc.com/live |
Resource Availablity
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dimiter Dimitrov (mit666666@pitt.edu).
Materials Availability
All requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact author. This includes antibodies, viruses, plasmids and proteins. All reagents will be made available on request after completion of a Material Transfer Agreement.
Data and Code Availability
Antibody nucleotide sequence has been deposited to GenBank with an accession number of GenBank: MT943599. The antibody is only allowed for non-commercial use. All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request.
Experimental Model and Subject Details
Cells and virus
Vero E6 (CRL-1586, American Type Culture Collection (ATCC)) and 293T (ATCC) were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, and 100 U/mL of penicillin–streptomycin. 293T stably expressing SARS-CoV-2 and human ACE2 was cultured in DMEM medium containing 200 μg/ml Zeocin. HEK293F and expi293F were cultured in FreeStyle 293 serum free medium (ThermoFisher, Cat#12338018) and Expi293 Expression Medium (ThermoFisher, Cat# A1435103), respectively. The SARS-CoV-2 spike pseudotyped HIV-1 backboned virus is packaged in 293T cells after transfecting pNL4-3.luc.RE and pcDNA3.1 S plasmids. The SARS-CoV-2 (US_WA-1/2020) and SARS-CoV2/Canada/ON/VIDO-01/2020 obtained from Centers for Disease Control and Prevention were propagated in Vero E6 cells. The recombinant SARS-CoV-2-SeattlenLuc virus and the mouse ACE2 adapted SAR-CoV-2 virus (carrying a Q498T/P499Y mutation in RBD) recovered by the reverse genetics was produced in VeroE6 cells. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 facilities using appropriate positive pressure air respirators and protective equipment.
Recombinant proteins
The recombinant proteins SARS-CoV-2 RBD-his, RBD mutants, RBD-Fc, ACE2-hFc were subcloned into pcDNA3.1 expression plasmids, and expressed in expi293F cells. Proteins with his tag were purified by Ni-NTA affinity chromatography and protein with Fc tag purified by protein A chromatography. Protein purity was estimated as > 95% by SDS-PAGE and protein concentration was measured spectrophotometrically (NanoVue, GE Healthcare).
Monoclonal antibodies
VH ab8 antibody was identified by panning of the phage library. VH-Fc ab8 were constructed by fusing VH to human IgG1 Fc with the native IgG1 hinge. IgG1 ab1 was obtained by our lab through panning of a Fab phage library. MERS-CoV-specific IgG1 m336 and SARS-CoV antibody IgG1 CR3022 sequences from other groups were subcloned into the pDR12 plasmid for expression. VH ab8 (in a phagemid pComb3x with a Flag tag) was expressed in HB2151 E. coli and purified by Ni-NTA affinity chromatography. All other IgG1 were expressed in expi293 cells and purified with protein A chromatography.
Mouse and hamster experiments
For the mouse model, BALB/c mice purchased from Envigo (BALB/cAnNHsd, stock# 047, immunocompetent, 11-12 months of age, female) were used for all experiments. They are drug/test naive and negative for pathogens. Animals were not involved in any previous studies. Animals were housed in groups of 5 animals per cage and fed standard chow diet. The study was carried out in accordance with the recommendations for care and use of animals by the Office of Laboratory Animal Welfare (OLAW), National Institutes of Health and the Institutional Animal Care. All mouse studies were performed at the University of North Carolina (Animal Welfare Assurance #A3410-01) using protocols (19-168) approved by the UNC Institutional Animal Care and Use Committee (IACUC) and all virus studies were performed in ABSL3 facilities at UNC. Virus inoculations were performed under anesthesia and all efforts were made to minimize animal suffering. For evaluating prophylactic efficacy of VH-Fc ab8, mice were intraperitoneally treated (12 hours before infection) with different doses of VH-Fc ab8 followed by intranasal challenge with 105 PFU of mouse-adapted SARS-CoV-2. Two days post infection, mice were sacrificed and perfused with 10 mL PBS. Then lung was harvested for viral titer as determined by the plaque assay. For the hamster model, studies were approved by the University Animal Care Committee (UACC) of the University of Saskatchewan according to the guidelines of the Canadian Council on Animal Care (CCAC). Hamsters were purchased from Charles River (male, immunocompetent, healthy, drug/test naive, free of pathogens). Hamsters were not involved in previous procedures. Hamsters are housed in microisolater cages, typically 3-7/cage. The cages have BioFresh bedding with Crinkle bedding added. Hamsters have access to food and water ab libitum. Food is Lab Diet 5P00 ProLab RMH300. Cages are changed weekly or as needed and spot cleaned. For experiment, hamsters were intraperitoneally treated with VH-Fc ab8 either 24 hr before (prophylaxis) or 6 hr (therapy) after intranasal challenge of 1 × 105 TCID50 of SARS-CoV-2. Nasal washes and oral swabs were collected at day 1, 3 and 5 post infection (dpi). Hamsters were bled at 1 and 5 dpi. All hamsters were euthanized on 5 dpi. At euthanasia, lungs were collected for RNA isolation. For viral titer determination, VeroE6 cells TCID50 assay was used. For testing viral RNA, viral RNA RT-qPCR was used. For testing antibody concentration at sera and lung, SARS-CoV-2 S1 ELISA was used. For histopathology, 10% formalin fixed and paraffin embedded tissues were processed with either hematoxylin and eosin stain (H&E) or immunohistochemistry (IHC). Lung lobes were scored based on pathology using microscopy.
Method Details
Generation, Expression and Characterization of SARS-CoV-2 RBD, S1-Fc, ACE2-Fc, IgG1 m336, and Fab CR3022
The SARS-CoV-2 S and the anti-SARS-CoV antibody IgG1 CR3022 and genes were synthesized by IDT (Coralville, Iowa). MERS-CoV-specific IgG1 m336 antibody was expressed in human mammalian cell as described previously (Ying et al., 2014a). Briefly, IgG1 m336 light chain and heavy chain Fd were subcloned into the pDR12 vector containing dual promoters and a IgG1 Fc cassette. The recombinant plasmid was sequenced and transfected into expi293 cells for expression. The human angiotensin converting enzyme 2 (ACE2) gene was ordered from OriGene (Rockville, MD). The RBD domain (residues 330-532) and S1 domain (residues 14-675) and ACE2 (residues 18-740) genes were cloned in frame to human IgG1 Fc in the mammalian cell expression plasmid pcDNA3.1. The RBD protein with an AviTag followed by a 6 × His tag at C-terminal was subcloned similarly. These proteins were expressed with Expi293 expression system (Thermo Fisher Scientific) and purified with protein A resin (GenScript) and by nickel- nitrilotriacetic acid (Ni-NTA) resin (Thermo Fisher Scientific). The Fab CR3022 antibody gene with a His tag was cloned into pCAT2 plasmid (developed in house) for expression in HB2151 bacteria and purified with Ni-NTA resin. Protein purity was estimated as > 95% by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and protein concentration was measured spectrophotometrically (NanoVue, GE Healthcare).
Generation of a human VH library, Selection of Binders and Conversion of VH to VH-Fc Fusion Protein
Unlike camel VHHs, which naturally evolved to be autonomously stable, human VH is usually unstable and easy to aggregate in the absence of VL (Li et al., 2016; Nguyen et al., 2000). However, human VH can be selected or engineered with high stability and solubility. To facilitate identification of stable VH binders, we chose engineered germline VH3-23 as our library scaffold (Chen et al., 2008b). Our human VH phage display library was made by grafting heavy chain CDR1, 2, 3 genes derived from 12 healthy donors’ peripheral blood monocytes (PBMCs) and splenocytes (Takara, Cat. No. 636525) into their cognate positions of a stable scaffold (based on the germline VH3-23) in a manner similar to the method we previously described but without mutagenesis of CDR1 (Chen et al., 2008a). Briefly, CDRs were PCR-amplified by using primers with degenerated adaptors covering CDRs edge regions from diverse VH families in one end, and with sequences annealing to the VH3-23 framework (FR) regions in the other end. The PCR products were then assembled by overlapping extension PCR by using primers with homologous ending. The whole VH was assembled by overlapping FR1-CDR1-FR2-CDR2 and FR3-CDR3-FR4 fragments. After assembly, the VH fragment was Sfi I digested followed by ligated into Sfi I linearized pComb3x phagemid. The recombinant phagemid was then purified, desalted and concentrated for electroporation of bacteria TG1, from which the VH phage particles were rescued and produced. The library size was determined by tittering transformants. The library quality (diversity) was checked by randomly Sanger sequencing hundreds of VH clones and also evaluated by panning of diverse antigens. This library contains very large number of clones (1011). For panning, the VH library was alternatively panned against biotinylated RBD-his and RBD-Fc proteins. RBD biotinylation occurred through biotin ligase (BirA) mediated enzymatic conjugation of a single biotin on AviTag (GLNDIFEAQKIEWHE) (Fairhead and Howarth, 2015). The panning was for 3 rounds with input antigens of 10 μg RBD-his, 2 μg RBD-Fc and 0.5 μg RBD-his for the 1st, 2nd and 3rd round, respectively. The panning process begun with incubation of antigens with 1012 VH phage particles followed by washing with phosphate-buffered saline (PBS) containing 0.1% Tween-20. Bound phage pulled down by streptavidin-M280-Dynabeads were rescued by log-phase TG1 cells with the M13KO7 helper phage. After the 3rd round panning, positive clones were selected by soluble expression monoclonal (SEM) ELISA followed by sequencing (Chen et al., 2008b). VH binders were further screened for their binding affinity, stability and ACE2 competition. For conversion to Fc-fusion, the VH gene was subcloned into pSecTag B vector containing human IgG1 Fc fragment. VH-Fc ab8 was expressed as described above.
Enzyme-Linked Immunosorbent Assays (ELISAs)
For detection of RBD biotinylation efficacy, horseradish peroxidase (HRP) conjugated streptavidin was used. For conformation of function of RBD-his after biotinylation, 100 ng ACE2-Fc was coated into the plates followed by addition of serially diluted biotinylated RBD-his. HRP conjugated streptavidin was used for detection. For other ELISAs, the SARS-CoV-2 RBD (residues 330-532) protein was coated on 96-well plates (Costar) at 100 ng/well in PBS overnight at 4°C. For screening SEM. ELISA, clones randomly picked from the infected TG1 cells were incubated with immobilized antigen. Bound phages were detected with HRP-conjugated mouse anti-FLAG tag Ab (Sigma-Aldrich). For the VH-Fc binding assay, HRP-conjugated goat anti-human IgG Fc (Sigma-Aldrich) was used for detection. For the competition ELISA with hACE2, 2 nM of human ACE2-mouse Fc was incubated with serially diluted VH, or VH-Fc, and the mixtures were added to RBD coated wells. After washing, bound ACE2-mouse Fc was detected by HRP-conjugated anti mouse IgG (Fc specific) (Sigma-Aldrich). For evaluation of ACE2 blocking of VH ab8 binding to RBD, 10 nM VH ab8 was incubated with coated RBD in the presence of various concentration of ACE2-His (Sino Biological), and the bound VH ab8 was detected by HRP conjugated anti FLAG antibody. For evaluation of conformational changes of the epitope mapping RBD mutants, we used a mouse polyclonal anti SARS-CoV-2 RBD antibody (Sino biological, Cat. No. 40592-MP01) and the human IgG1 CR3022 antibody. For measuring the binding of VH-Fc ab8 to RBD mutants, 100 ng RBD mutant was coated on 96-wells plates and incubated with VH-Fc ab8 with binding detected by using HRP conjugated anti human Fc antibody. To evaluate the binding of VH-Fc ab8 and IgG1 ab1 to human FcγRs, recombinant human FcγRIA, IIA, IIIA were coated on 96-wells plates followed by addition of biotinylated VH-Fc ab8 and IgG1 ab1. Binding was detected by the streptavidin-HRP. All colors were developed by 3,3′,5,5′-tetramethylbenzidine (TMB, Sigma) and stopped by 1 M H2SO4 followed by recording absorbance at 450 nm. Experiments were performed in duplicate and the error bars denote ± 1 SD.
BLItz
Antibody affinities and avidities were analyzed by the biolayer interferometry BLItz (ForteBio, Menlo Park, CA). For measuring VH ab8 affinity, the RBD-Fc was mounted on the protein A sensor (ForteBio: 18-5010). 125 nM, 250 nM and 500 nM VH ab8 were used for association. For measuring avidity of VH-Fc ab8, biotinylated RBD-Fc was immobilized on streptavidin biosensors (ForteBio: 18–5019) for 2 min and equilibrated with Dulbecco’s phosphate-buffered saline (DPBS) (pH = 7.4) to establish baselines. 50 nM, 100 nM and 200 nM VH-Fc ab8 were chosen for association. The association was monitored for 2 min and then the antibody was allowed to dissociate in DPBS for 4 min. The ka and kd were derived from sensorgrams fittings and used for Kd calculation. For the competitive Blitz, 500 nM VH-Fc ab8 was loaded onto the RBD-Fc coated sensor for 300 s to reach saturation followed by dipping the sensor into a 100 nM ACE2-Fc or Fab CR3022 solution in the presence of 500 nM VH-Fc ab8. The association was monitored for 300 s. The signals from 100 nM hACE2 or CR3022 binding to the RBD-Fc coated sensor in the absence of VH-Fc ab8 was independently recorded in parallel. Competition was determined by the percentage of signal in the presence of VH-Fc ab8 to signal in the absence of VH-Fc ab8 (< 0.7 is considered to be competitive) (Wu et al., 2020a).
SARS-CoV-2 RBD Mutants and Epitope Mapping by Ala Scanning
RBD mutants, N354D, N354D/D364Y, V367F, R408I, W436R were purchased from Acro Biosystems. F342L and K458R were bought from Sino Biological. RBD mutants G476S and V483A, plus the alanine (Ala) scanning mutants N439A, G446L, L455A, F456A, A475I, F486A, Q493A, Q498A, N501A, Y505A were constructed by site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, cat. no. 200521). Mutants were expressed and purified according to the abovementioned RBD purification procedures. ELISA was used to evaluate the binding of these mutants compared to the wild-type RBD.
Electron Microscopy for SARS-CoV-2 S Trimer Complexed with VH ab8
A. Expression and Purification
The codon optimized SARS-CoV-2 2P S protein ectodomain construct (GenBank: YP_009724390.1) was C-terminally tagged with 8xHis and a twin Strep tag and cloned into the mammalian expression vector pcDNA 3.1 (Synbio). HEK293F cells were grown in suspension culture using FreeStyle media (ThermoFisher) at 37°C in a humidified CO2 incubator (8% CO2). Cells were transiently transfected at a density of 1 × 106 cells/ml using branched polyethylenimine (PEI) (Sigma) (Portolano et al., 2014). Media was exchanged after 24 h and supplemented with 2.2 mM valproic acid. Supernatant was harvested by centrifugation after 4 days, filtered and loaded onto a 5 mL HisTrap HP column (Cytiva). The column was washed with buffer (20 mM Tris pH 8.0, 500 mM NaCl, 20 mM imidazole) and the protein was eluted with buffer (20 mM Tris pH 8.0, 500 mM NaCl, 500 mM imidazole). Purified protein was concentrated (Amicon Ultra 100 kDa cut off, Millipore Sigma) and loaded onto a Superose 6 column (Cytiva) equilibrated with GF buffer (20 mM Tris pH 8.0 and 150 mM NaCl). Peak fractions were pooled and concentrated to 1.3 mg/ml (Amicon Ultra 100 kDa cut off, Millipore Sigma).
B. Electron Microscopy Specimen Preparation and Data Collection
Purified S protein ectodomain (0.04 mg/ml) was mixed with VH ab8 (0.02 mg/ml) or soluble ACE2 (0.02 mg/mL) and incubated on ice for 10 mins. For the competition experiment, the S protein (0.04 mg/ml) was first incubated on ice with VH ab8 (0.02 mg/ml) for 10 mins then followed by addition of ACE2 (0.02 mg/mL) for another 10 mins. The mixtures (4.8 μl) were applied to 300-mesh copper grids coated with continuous ultrathin carbon. Grids were plasma cleaned using an H2/O2 gas mixture for 15 s in a Solarus plasma cleaner (Gatan Inc.) prior to adding the sample. Samples were allowed to adsorb for 30 s before blotting away excess liquid, followed by a brief wash with MilliQ H2O. Grids were stained by three successive applications of 2% (w/v) uranyl formate (20 s, 20 s, 60 s). Grids containing S protein ectodomain with VH ab8, and S protein ectodomain mixed with both VH ab8 and soluble ACE2 were imaged using a 200 kV Glacios transmission electron microscope (ThermoFisher Scientific) equipped with a Falcon3 camera operated in linear mode. Using EPU automated acquisition software (ThermoFisher Scientific), 15-frame movies were collected at 92,000x magnification (corresponding to a physical pixel size of 1.6 Å) over a defocus range of −0.5 to −3.0 μm with an accumulated total dose of 40 e-/Å2/movie. Grids containing purified S protein ectodomain (0.04 mg/mL) with soluble ACE2 (0.02 mg/mL) were imaged using a 200kV Glacios transmission electron microscope equipped with a Ceta 16M CMOS camera (ThermoFisher Scientific). Micrographs were collected at 92,000x magnification (physical pixel 1.6 Å) over a defocus range of −0.5 to −3.0 μm with a total dose of 50 e-/Å2 using EPU automated acquisition software.
C. Image Processing
Motion correction and CTF estimation were performed in RELION (3.1) (Scheres, 2012). Particles were picked by crYOLO (1.7.4) (Wagner et al., 2019) with pre-trained model for negative stain data. After extraction, particles were imported to cryoSPARC live (v2.15.1) (Punjani et al., 2017) and subjected to 2D classification and 3D heterogeneous classification. Final density maps were obtained by 3D homogeneous refinement. Figures were prepared using UCSF Chimera (Pettersen et al., 2004).
Flow Cytometry Analysis (FACS)
Full-length S protein of SARS-CoV-2 with native signal peptide replaced by the CD5 signal peptide were codon-optimized and synthesized by IDT. The S gene was subcloned into our in-house mammalian cell expression plasmid, which were used to transiently transfect 293T cells cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% FBS, 1% penicillin-streptomycin (P/S). The comparisons of ACE2-Fc, IgG1 CR3022 and VH-Fc ab8 binding to both blank 293T and 293T overexpressing S (293T-S) were performed. For the determination of binding avidity of VH-Fc ab8 and ACE2-Fc to the cell surface S, serially diluted antibodies or ACE2-Fc with highest concentration of 1 μM were incubated with cells, and after washing, bound antibodies were detected by phycoerythrin (PE) conjugated anti-human Fc antibody (Sigma-Aldrich). PE-A+ cells were detected by flow cytometry using BD LSR II (San Jose, CA). The gating of PE-A+ population was performed by the FlowJo software, which was plotted against the concentrations of proteins to calculate FC50 by non-linear fitting in Graphpad Prism 7 (San Diego, CA). To evaluate ACE2 blocking of VH ab8 binding to cell surface associated S, gradient concentrations of ACE2-his in the presence of 1 μM VH ab8 (Flag tag) were incubated with 293-S cells. After washing, VH ab8 binding was detected by PE conjugated anti FLAG tag antibody.
Cell-Cell Fusion Inhibition Assay
To test antibody mediated inhibition of cell fusion, the β-galactosidase (β-gal) reporter gene based quantitative cell fusion assay was used (Xiao et al., 2003). In this assay, 293T-S cell expression of T7 RNA polymerase was achieved by infection with vaccinia virus VTF7.3, while 293T-ACE2 cell expression of T7 promoter controlled β-Gal was obtained by infection with vaccinia virus VCB21R. β-Gal will be expressed only after fusion of the two types of cells, which can be monitored by chromogenic reactions using β-Gal substrate. To assay cell-cell fusion, 293T cells stably expressing SARS-CoV-2 S (293T-S) cells were infected with T7 polymerase-expressing vaccinia virus (vTF7-3), and 293T cells stably expressing ACE2 (293T-ACE2) were infected with vaccinia virus (vCB21R Lac-Z) encoding T7 promotor controlled β-gal. Two hours after infection, cells were incubated with fresh medium and transferred to 37°C for overnight incubation. The next day, 293T-S cells were pre-mixed with serially diluted antibodies or ACE2-Fc at 37°C for 1 h followed by incubation with 293T-ACE2 cells at a 1:1 ratio for 3 h at 37°C. Then cells were then lysed, and the β-gal activity was measured using β-galactosidase assay kit (substrate CPRG, G-Biosciences, St. Louis, MO) following the manufacturer’s protocol. Fusion inhibition percentage (sample reading, F) was normalized by maximal fusion (reading, Fmax) of 293T-S and 293T-ACE2 cells in the absence of antibodies using this formula: Fusion inhibition % = [(Fmax-F)/(Fmax - Fblank)] × 100%, in which Fblank refers to the OD reading of 293T-S and 293T incubation wells. Fusion inhibition percentage was plotted against antibody concentrations. Experiments were performed in duplicate and the error bars denote ± 1 SD.
Pseudovirus Neutralization Assay
Pseudovirus neutralization assay was performed based on previous protocols (Zhao et al., 2013). Briefly, HIV-1 backbone based pseudovirus was produced in 293T cells by co-transfection with plasmid encoding SARS-CoV-2 S protein and plasmid encoding luciferase expressing HIV-1 genome (pNL4-3.luc.RE) using PEI. Pseudovirus-containing supernatants were collected 48 h later and concentrated using Lenti-X concentrator kit (Takara, CA). Pseudovirus neutralization assay was then performed by incubation of SARS-CoV-2 pseudovirus with serially diluted antibodies or ACE2-Fc for 1 h at 37°C, followed by addition of the mixture into pre-seeded 293T-ACE2 cells. The mixture was then centrifuged at 1000 × g for 1 hour at room temperature. The medium was replaced 4 hr later. After 24 h, luciferase expression was determined by Bright-Glo kits (Promega, Madison, WI) using BioTek synergy multi-mode reader (Winooski, VT). Cells only and virus only wells were included and used for normalization. The 50% pseudovirus neutralizing antibody titer (IC50) was calculated using Graphpad Prism 7. Experiments were performed in duplicate and the error bars denote ± 1 SD.
SARS-CoV and SARS-CoV-2 Microneutralization Assay
The standard live virus-based microneutralization (MN) assay was used as previously described (Agrawal et al., 2016a, 2016b; Du et al., 2013, 2014). Briefly, serially three-fold and duplicate dilutions of individual monoclonal antibodies (mAbs) were incubated with 120 pfu of SARS-CoV or SARS-CoV-2 at room temperature for 2 h before transferring into designated wells of confluent Vero E6 cells grown in 96-well microtiter plates. Vero E6 cells cultured with medium with or without virus were included as positive and negative controls, respectively. MERS-CoV RBD-specific neutralizing m336 mAb (Ying et al., 2014a) were used as additional controls. After incubation at 37°C for 4 days, individual wells were observed under the microcopy for the status of virus-induced formation of cytopathic effect. The efficacy of individual mAbs was expressed as the lowest concentration capable of completely preventing virus-induced cytopathic effect in 100% of the wells.
SARS-CoV and SARS-CoV-2 Reporter Gene Neutralization Assay
Full-length viruses expressing luciferase were designed and recovered via reverse genetics as described previously (Scobey et al., 2013; Yount et al., 2003). Briefly, the SARS-CoV-2 RNA from infected cell culture was reverse-transcribled and constructed into the seven contiguous genomic cDNA subclones with interconnecting junctions, which were then BsaI/BsmBI digested and ligated into a full-length SARS-CoV-2 genome cDNA through the cohesive ends. A silent mutation of T15102A was introduced into a conserved region in nsp12 to differentiate our recombinant viruses from the circulating SARS-CoV-2 strains through Sanger sequencing. The reporter viruse was synthesized by replacing a 276-bp region in ORF7 with a GFP-fused nanoluciferase (nLuc) gene. After assembly into full-length cDNA, full-length RNA was in vitro transcribed and was electroporated into Vero E6 cells. Virus stocks were propagated on Vero E6 cells in minimal essential medium containing 10% fetal bovine serum (HyClone) and supplemented with penicillin/kanamycin (Gibico). Viruses were tittered in Vero E6 USAMRID cells to obtain a relative light units (RLU) signal of at least 20 × the cell only control background. Ab or ACE2-Fc were serially diluted 4-fold up to eight dilution spots with at a starting dilution 100 μg/ml, and were incubated with SARS-CoV-UrbaninLuc and SARS-CoV-2-SeattlenLuc viruses at 37°C with 5% CO2 for 1 hour. Then virus-antibody dilution complexes were added to the pre-seeding E6 USAMRID cells (20,000) in duplicate. Virus-only controls and cell-only controls were included in each neutralization assay plate. Following infection, plates were incubated at 37°C with 5% CO2 for 48 hours. Then cells were lysed and luciferase activity was measured via Nano-Glo Luciferase Assay System (Promega) according to the manufacturer specifications. SARS-CoV and SARS-CoV-2 neutralization IC50 were defined as the sample concentration at which a 50% reduction in RLU was observed relative to the average of the virus control wells. Experiments were performed in duplicate and IC50 was obtained by the non-linear fitting of neutralization curves in Graphpad Prism 7.
Evaluation of the VH-Fc ab8 Protective Efficacy in a Mouse Adapted SARS-CoV-2 Model
A recombinant mouse ACE2 adapt SARS-CoV-2 variant was constructed by introduction of two amino acid changes (Q498T/P499Y) at the ACE2 binding pocket in RBD. Virus stocks were grown on Vero E6 cells and viral titer was determined by plaque assay (Dinnon et al., 2020). Groups of 5 each of 10 to 12-month old female BALB/c mice (Envigo, #047) were treated prophylactically (12 hours before infection) by intraperitoneal injection with 36, 8, or 2 mg/kg of VH-Fc ab8, respectively. Mice were challenged intranasally with 105 PFU of mouse-adapted SARS-CoV-2. Two days post infection, mice were sacrificed and lung viral titer was determined by the plaque assay. To exclude the residual lung antibody impact on viral titration, mice were euthanized and perfused with 10 mL of PBS via cardiac puncture before lung harvest for viral titration. For virus titration, the caudal lobe of the right lung was homogenized in PBS. The resulting homogenate was serial-diluted and inoculated onto confluent monolayers of Vero E6 cells, followed by agarose overlay. Plaques were visualized via staining with Neutral Red on day 2 post infection. To measure the viral RNA in the lung, tissue homogenate lysed in Trizol LS (Thermofischer) was then processed with Thermofischer Trizol RNA isolation protocol followed by RT-qPCR using the QuantiFast Probe RT-PCR kit (QIAGEN) to amplify a portion of upE gene. The 50% tissue culture infectious doses (TCID50) equivalence were estimated by running serial dilutions of known TCID50 standards.
Evaluation of the VH-Fc ab8 Prophylactic and Therapeutic Efficacy in a Hamster Model of SARS-CoV-2 Infection
SARS-CoV2/Canada/ON/VIDO-01/2020 was propagated on Vero’76 cells using DMEM with 2% FBS and 1 μg/ml L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TCPK) trypsin. Infectious work with SARS-CoV-2 was approved by the Biosafety Protocol Approval Committee (BPAC) at the University of Saskatchewan and performed in the high containment laboratories at VIDO-InterVac. Male hamsters (9-week-old) were obtained from Charles River (Montreal, QC). For evaluations of prophylactic efficacy, all hamsters (n = 7) were injected intraperitoneally with 10 mg/kg of VH-Fc ab8 24 hours prior to intranasal challenge of 50 μl/nare containing a total of 1 × 105 TCID50 of SARS-CoV-2. For the therapeutic group, hamsters were infected as above and treated intraperitoneally with 10 mg/kg (n = 3) or 3 mg/kg (n = 4) of VH-Fc ab8 6 hours post-infection. Untreated hamsters were kept as a control. Nasal washes and oral swabs were collected at day 1, 3 and 5 post infection (dpi). Hamsters were bled at 1 and 5 dpi. All hamsters were euthanized on 5 dpi. At euthanasia, lung lobes were collected for virus titration and RNA isolation. For viral titer determination, nasal washes were diluted in a 10-fold dilution series and absorbed on Vero’76 cells in triplicates for 1 hour at 37°C. Inoculum was removed and replaced with fresh DMEM containing 2% FBS, penn/strep and 1μg/ml TPCK. Cytopathic effect was scored on day 3 and day 5 post infection. The limit of detection is 13.6 TCID50. For testing viral RNA, viral RNA isolated from nasal and oral swabs using the QiaAmp Viral RNA mini kit (QIAGEN) and the QuantiFast Probe RT-PCR kit (QIAGEN) to amplify a portion of upE gene. For RNA levels in tissues, 30 mg of tissue homogenate in buffer RLT were processed with the RNeasy kit (QIAGEN) followed by RT-qPCR as above. TCID50 equivalence were estimated by running serial dilutions of known TCID50 standards. For testing Ab8 concentrations post injection at hamster sera and lung tissue, SARS-CoV-2 spike-1 ELISA was used. S1 protein was coated at 1 μg/ml overnight at 4°C in PBS onto MaxiSorp plates (Nunc). The following day plates were blocked with 5% skim milk and 0.05%Tween20. Serum collected on day 1 and day 5 post-challenge was diluted 1:100 and absorbed for 1 hour at 37°C. Plates were washed and goat anti human IgG-HRP was added. Plates were washed and subsequently developed with OPD (o-phenylenediamine dihydrochloride) substrate. Optical density was measured at 450 nm after 30 mins of incubation. For lung tissues, after blocking homogenates were diluted 1:10 and absorbed overnight at 4°C followed by detection with anti-human IgG-HRP and substrate as stated above. The control hamster lung homogenate was used for background correction. For histopathology on day 5 p.i., 10% formalin fixed and paraffin embedded tissues were processed with either hematoxylin and eosin stain (H&E) or immunohistochemistry (IHC) for detection of SARS-CoV2 antigen; in IHC after blocking tissue slides were treated with anti-nucleocapsid rabbit polyclonal antibodies followed with anti-rabbit HRP antibody.
Dynamic Light Scattering (DLS)
For evaluation of aggregation propensity, VH ab8 and VH-Fc ab8 were buffer-changed to DPBS and filtered through a 0.22 μm filter. The concentration was adjusted to 4 mg/mL; 500 μL samples were incubated at 37°C. On day 0, day 1 and day 6, samples were taken out for DLS measurement on Zetasizer Nano ZS ZEN3600 (Malvern Instruments Limited, Westborough, MA) to determine the size distributions of protein particles.
Size Exclusion Chromatography (SEC)
The Superdex 200 Increase 10/300 GL chromatography (GE Healthcare, Cat. No. 28990944) was used. The column was calibrated with protein molecular mass standards of Ferritin (Mr 440 000 kDa), Aldolase (Mr 158 000 kDa), Conalbumin (Mr 75 000 kDa), Ovalbumin (Mr 44 000 kDa), Carbonic anhydrase (Mr 29 000 kDa), Ribonuclease A (Mr 13 700 kDa). 150 μl filtered proteins (1.5 mg/ml) in PBS were used for analysis. Protein was eluted by DPBS buffer at a flow rate of 0.5 ml/min.
Membrane Proteome Array Assay
Integral Molecular, Inc. (Philadelphia, PA) performed specificity testing of VH-Fc ab8 using the Membrane Proteome Array (MPA) platform. The MPA comprises 5,300 different human membrane protein clones, each overexpressed in live cells from expression plasmids that are individually transfected in separate wells of a 384-well plate (Tucker et al., 2018). The entire library of plasmids is arrayed in duplicate in a matrix format and transfected into HEK293T cells, followed by incubation for 36 h to allow protein expression. Before specificity testing, optimal antibody concentrations for screening were determined by using cells expressing positive (membrane-tethered Protein A) and negative (mock-transfected) binding controls, followed by flow cytometric detection with an Alexa Fluor-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Based on the assay setup results, VH-Fc ab8 (20 μg/ml) was added to the MPA. Binding across the protein library was measured on an iQue3 (Ann Arbor, MI) using the same fluorescently labeled secondary antibody. To ensure data validity, each array plate contained positive (Fc-binding; SARS-CoV-2 S protein) and negative (empty vector) controls. Identified targets were confirmed in a second flow cytometric experiment by using serial dilutions of the test antibody. The identity of each target was also confirmed by sequencing.
Quantification and Statistical Analysis
For the mouse model, the statistical significance of difference between VH-Fc ab8 treated and control mice lung virus titers was determined by the two-tailed, unpaired, Student’s t test calculated using GraphPad Prism 7.0. A p value < 0.05 was considered significant. ∗∗ p < 0.01. For the mice lung viral titer after perfusion, viral RNA and hamster lung viral RNA, statistical significance was determined by the Mann-Whitney U test. A p value < 0.05 was considered significant. ns: p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. For comparing VH-Fc ab8 and IgG1 ab1 concentration, significance analysis was determined by the two-way ANOVA followed by Tukey test in GraphPad Prism 7.0. A p value < 0.05 was considered significant. ns: p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We would like to thank the members of our group, Dontcho Jelev, Megan Shi, Cynthia Adams, Du-San Baek, Ye-Jin Kim, and Xiaojie Chu for their helpful discussions. We thank Dr. Kevin McCormick from the University of Pittsburgh, Rui Gong from the Institute of Virology in Wuhan, and Rachel Fong from Integral Molecular for helpful suggestions. We would also like to thank Jocelyne Lew and Vinoth Manoharan for technical assistance and the members of the Clinical Research and Animal Care team at VIDO-InterVac, as well as Yanyun Huang and Dale Godson (Prairie Diagnostic Services Inc.). This work was supported by the University of Pittsburgh Medical Center. D.R.M. is funded by NIH (F32 AI152296), a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award, and NIH NIAID (T32 AI007151). R.S.B. is supported by NIH (AI132178 and AI108197). Work in the Subramaniam laboratory is supported by a Canada Excellence Research Chair Award and a grant from Genome BC, Canada. Some monoclonal antibodies were generated by the UNC Protein Expression and Purification (PEP) core facility, which is funded by NIH (P30CA016086).
Author Contributions
D.S.D., R.S.B., C.-T.T., J.W.M., S.S., D.F., and W.L. conceived and designed the research. W.L. identified and characterized antibodies. X.L. and Z.S. helped to make libraries, characterized antibodies, and performed the cell fusion pseudovirus assays. C.C. made the RBD and ACE2. L.Z. made and characterized reagents. M.L.U. and E.C.P. characterized proteins and helped with the proteome assay. D.M. and A.D. performed the live virus neutralization assays. A.S., S.S.K., D.F., and S.L. performed the animal studies. A.B., S.C., K.L., D.M., S.S.S., X.Z., and S.S. produced and purified the S trimer, carried out the EM experiments, and analyzed the structure-related results. D.S.D. and W.L. wrote the first draft of the article. All authors discussed the results and contributed to the manuscript.
Declaration of Interests
W.L., C.C., Z.S., J.W.M., and D.S.D. are co-inventors of a patent, filed on March 12 by the University of Pittsburgh, related to ab8 described in this paper.
Published: September 4, 2020
References
- Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.H., Couch R.B., Tseng C.T. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccin. Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A.S., Ying T., Tao X., Garron T., Algaissi A., Wang Y., Wang L., Peng B.H., Jiang S., Dimitrov D.S., Tseng C.T. Passive Transfer of A Germline-like Neutralizing Human Monoclonal Antibody Protects Transgenic Mice Against Lethal Middle East Respiratory Syndrome Coronavirus Infection. Sci. Rep. 2016;6:31629. doi: 10.1038/srep31629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.06.021. Published online July 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu Z., Feng Y., Dimitrov D.S. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. USA. 2008;105:17121–17126. doi: 10.1073/pnas.0805297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu Z., Feng Y., Xiao X., Dimitrov D.S. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J. Mol. Biol. 2008;382:779–789. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Liu X., Wang C., Zhang X., Ren L., Jin Q., Wang J., Yang W. Humanized Single Domain Antibodies Neutralize SARS-CoV-2 by Targeting Spike Receptor Binding Domain. bioRxiv. 2020 doi: 10.1101/2020.04.14.042010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang G.-Y., Zhang B., McKee K., O’Dell S., Kwon Y.D., Zhou T., Blinn J., Lloyd K., Parks R., Von Holle T. Eliminating antibody polyreactivity through addition of N-linked glycosylation. Protein Sci. 2015;24:1019–1030. doi: 10.1002/pro.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K. Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 2015;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. bioRxiv. 2020 doi: 10.1101/2020.05.06.081497. [DOI] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.-T.K., Zhou Y., Jiang S. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS ONE. 2013;8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z., Tao X., Yu H., Sun S., Tseng C.T. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M., Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freise A.C., Wu A.M. In vivo imaging with antibodies and engineered fragments. Mol. Immunol. 2015;67(2 Pt A):142–152. doi: 10.1016/j.molimm.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen M.M., De Haard H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhu Q., Liu S., Zhou Y., Yang B., Li J., Jiang S. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology. 2005;334:74–82. doi: 10.1016/j.virol.2005.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020 doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R.K. Physiological Barriers to Delivery of Monoclonal Antibodies and Other Macromolecules in Tumors. Cancer Res. 1990;50:814s–819s. [PubMed] [Google Scholar]
- Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg. Microbes Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. Growth kinetics of SARS-coronavirus in Vero E6 cells. Biochem. Biophys. Res. Commun. 2005;329:1147–1151. doi: 10.1016/j.bbrc.2005.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse P.J., Sattentau Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- Lagassé H.A.D., Hengel H., Golding B., Sauna Z.E. Fc-Fusion Drugs Have FcγR/C1q Binding and Signaling Properties That May Affect Their Immunogenicity. AAPS J. 2019;21:62. doi: 10.1208/s12248-019-0336-8. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11:2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Prabakaran P., Chen W., Zhu Z., Feng Y., Dimitrov D.S. Antibody Aggregation: Insights from Sequence and Structure. Antibodies (Basel) 2016;5:19. doi: 10.3390/antib5030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Drelich A., Martinez D.R., Gralinski L., Chen C., Sun Z., Liu X., Zhelev D., Zhang L., Peterson E.C. Potent neutralization of SARS-CoV-2 in vitro and in an animal model by a human monoclonal antibody. bioRxiv. 2020 doi: 10.1101/2020.05.13.093088. [DOI] [Google Scholar]
- Li X., Song Y., Wong G., Cui J. Bat origin of a new human coronavirus: there and back again. Sci. China Life Sci. 2020;63:461–462. doi: 10.1007/s11427-020-1645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gao F., Gou L., Chen Y., Gu Y., Ao L., Shen H., Hu Z., Guo X., Gao W. Neutralizing Antibodies Isolated by a site-directed Screening have Potent Protection on SARS-CoV-2 Infection. bioRxiv. 2020 doi: 10.1101/2020.05.03.074914. [DOI] [Google Scholar]
- Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020 doi: 10.1126/science.abc5881. eabc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson A.L. Antibody fragments: hope and hype. MAbs. 2010;2:77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.K., Hamers R., Wyns L., Muyldermans S. Camel heavy-chain antibodies: diverse germline V(H)H and specific mechanisms enlarge the antigen-binding repertoire. EMBO J. 2000;19:921–930. doi: 10.1093/emboj/19.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilvebrant J., Tessier P.M., Sidhu S.S. Engineered Autonomous Human Variable Domains. Curr. Pharm. Des. 2016;22:6527–6537. doi: 10.2174/1381612822666160921143011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin M., Naranjo-Gomez M., Piechaczyk M. Antiviral Monoclonal Antibodies: Can They Be More Than Simple Neutralizing Agents? Trends Microbiol. 2015;23:653–665. doi: 10.1016/j.tim.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Portolano N., Watson P.J., Fairall L., Millard C.J., Milano C.P., Song Y., Cowley S.M., Schwabe J.W. Recombinant protein expression for structural biology in HEK 293F suspension cells: a novel and accessible approach. J. Vis. Exp. 2014;(92):e51897. doi: 10.3791/51897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka S., Ranabir M., Sourabrata C., Amit K.S., Mahitosh M., Siddik S. Mutations in Spike Protein of SARS-CoV-2 Modulate Receptor Binding, Membrane Fusion and Immunogenicity: An Insight into Viral Tropism and Pathogenesis of COVID-19. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12320567.v1. [DOI] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., Parcells M.S., Luo G., Li W., Zhong G. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-t., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020:eabc7520. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Rodríguez Y., Monsalve D.M., Acosta-Ampudia Y., Camacho B., Gallo J.E., Rojas-Villarraga A., Ramírez-Santana C., Díaz-Coronado J.C., Manrique R. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020;19:102554. doi: 10.1016/j.autrev.2020.102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J., McKenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Chen C., Li W., Martinez D.R., Drelich A., Baek D.-S., Liu X., Mellors J.W., Tseng C.-T., Baric R.S., Dimitrov D.S. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. MAbs. 2020;12:1778435. doi: 10.1080/19420862.2020.1778435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker D.F., Sullivan J.T., Mattia K.A., Fisher C.R., Barnes T., Mabila M.N., Wilf R., Sulli C., Pitts M., Payne R.J. Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proc. Natl. Acad. Sci. USA. 2018;115:E4990–E4999. doi: 10.1073/pnas.1716788115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverdorben F., Richter F., Hutt M., Seifert O., Malinge P., Fischer N., Kontermann R.E. Pharmacokinetic properties of IgG and various Fc fusion proteins in mice. MAbs. 2016;8:120–128. doi: 10.1080/19420862.2015.1113360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt M.J., Heijnen I.A., Capel P.J., Park S.Y., Ra C., Saito T., Verbeek J.S., van de Winkel J.G. FcR gamma-chain is essential for both surface expression and function of human Fc gamma RI (CD64) in vivo. Blood. 1996;87:3593–3599. [PubMed] [Google Scholar]
- Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., VIB-CMB COVID-19 Response Team Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L., Liu Q., Wang L., Feng Y., Wang Y. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Feng Y., Wang Y., Chen W., Dimitrov D.S. Monomeric IgG1 Fc molecules displaying unique Fc receptor interactions that are exploitable to treat inflammation-mediated diseases. MAbs. 2014;6:1201–1210. doi: 10.4161/mabs.29835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Du L., Ma C., Li Y., Li L., Poon V.K., Wang L., Yu F., Zheng B.J., Jiang S., Zhou Y. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol. J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Antibody nucleotide sequence has been deposited to GenBank with an accession number of GenBank: MT943599. The antibody is only allowed for non-commercial use. All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request.