Abstract
A prominent clinical symptom of 2019-novel coronavirus (nCoV) infection is hyposmia/anosmia (decrease or loss of sense of smell), along with general symptoms such as fatigue, shortness of breath, fever and cough. The identity of the cell lineages that underpin the infection-associated loss of olfaction could be critical for the clinical management of 2019-nCoV-infected individuals. Recent research has confirmed the role of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) as key host-specific cellular moieties responsible for the cellular entry of the virus. Accordingly, the ongoing medical examinations and the autopsy reports of the deceased individuals indicate that organs/tissues with high expression levels of ACE2, TMPRSS2 and other putative viral entry-associated genes are most vulnerable to the infection. We studied if anosmia in 2019-nCoV-infected individuals can be explained by the expression patterns associated with these host-specific moieties across the known olfactory epithelial cell types, identified from a recently published single-cell expression study. Our findings underscore selective expression of these viral entry-associated genes in a subset of sustentacular cells (SUSs), Bowman’s gland cells (BGCs) and stem cells of the olfactory epithelium. Co-expression analysis of ACE2 and TMPRSS2 and protein–protein interaction among the host and viral proteins elected regulatory cytoskeleton protein-enriched SUSs as the most vulnerable cell type of the olfactory epithelium. Furthermore, expression, structural and docking analyses of ACE2 revealed the potential risk of olfactory dysfunction in four additional mammalian species, revealing an evolutionarily conserved infection susceptibility. In summary, our findings provide a plausible cellular basis for the loss of smell in 2019-nCoV-infected patients.
Keywords: smell, COVID-19, pandemic, SARS-CoV-2, olfaction, olfactory sensory neurons (OSNs)
Introduction
The recent outbreak of the 2019-novel coronavirus (nCoV) triggered the need for community-scale deployment of diagnostic tests [1–4]. To date, various workgroups have extensively generated and analyzed the molecular profiles of the virus and the hosts [5, 6]. Some major efforts include isolation and sequencing of the viral genome from the airway epithelial cells [5–7]. Comparative genomics revealed that 2019-nCoV is closely related to the bat severe acute respiratory syndrome (SARS)-like coronaviruses (bat-SL-CoVZC45 and bat-SLCoVZXC21) [5]. Notably, the external subdomain of the 2019-nCoV spike, receptor-binding domain (RBD), shares ~40% identity at the amino acid level with other SARS-related coronaviruses [8]. Of note, most of the amino acid differences of the RBD are located in the external subdomain, which is responsible for the direct interaction of the virus with the host receptors [8]. Further, some of the recent reports underscore the role of angiotensin-converting enzyme 2 (ACE2) as a prominent surface receptor for the cellular entry of 2019-nCoV [9, 10]. A recent structural study involving the cryo-electron microscopy (EM) unraveled the strong interaction between the full-length viral spike protein and the human ACE2 receptor [11, 12]. Comparative analysis with the SARS-CoV strain further revealed that the intensity of the molecular interaction between viral spike and ACE2 is at least 10 times stronger in the case of the 2019-nCoV strain [12]. Notably, in addition to the ACE2-mediated attachment to the host cell surface, a successful viral entry also requires priming of the S protein via cellular proteases [13]. Priming involves cleavage of the S protein at the S1/S2 and the S2 sites, which, in turn, assists in the fusion of 2019-nCoV with the host cell membrane [14]. Notably, while leveraging antibody-mediated interventions, a recent report traced a novel entry route of 2019-nCoV into the host cell via CD147/BSG receptor [15]. Similarly, in addition to the TMPRSS2, other host cell proteases, such as cathepsin, are suggested to be involved in priming [16]. All these studies collectively reinforce the involvement of the aforementioned host proteins in successful 2019-nCoV entry into the host cell (Figure 1A). In order to determine the tissue- or organ-level impact of 2019-nCoV, various groups have traced the expression of these genes across organ/cell types [17–20]. Of note, many of these studies have leveraged single-cell expression studies to pinpoint the vulnerable cell subpopulation. High expression levels of these genes have been observed in a wide range of tissue/cell types such as epithelial cells of the esophagus, absorptive enterocytes of the intestines, mucosal cells of the oral cavity, proximal tubule cells of the kidney, myocardial cells of the heart, endothelial cells of the blood vessels, urothelial cells of the bladder, etc., thereby making them potentially vulnerable to the 2019-nCoV infection [17–21]. All these molecular findings are largely in line with the clinical symptoms reported worldwide, in which multi-organ failure is emerging as a major contributor to the infection-associated mortality [22]. While the loss of smell and taste has frequently been implicated to 2019-nCoV infection [23–25], its cellular basis has remained largely unexplored. The olfactory epithelium includes several distinct cell types, namely, horizontal basal cells (HBCs), microvillar cells (MVCs), Bowman’s gland cells (BGCs), globular basal cells (GBCs), olfactory ensheathing glia (OEGs), sustentacular cells (SUSs), immature olfactory sensory neurons and mature olfactory sensory neurons (iOSNs and mOSNs, respectively) [26]. Among these, OSNs are the key cell types that possess the receptors for odorant detection [26, 27]. In humans, there are at least ~400 functionally distinct OSNs [28]. In addition to these, the olfactory epithelium also possesses several other cell types for the maintenance of tissue architecture and homeostasis. SUS and MVC subtypes are responsible for providing metabolic as well as physical support to the olfactory epithelium [29]. GBCs and HBCs collectively constitute the basal stem cell population and mainly reside near the basal lamina [30]. These cell types ensure the renewal of the distinct cell types of the olfactory epithelium [31–33]. While the modes of action of the aforementioned cell types in mediating optimal olfactory function are already known, further investigation is needed to identify the specific cell types, in which the vulnerability to 2019-nCoV is largely unknown. Here we investigated the vulnerability of the olfactory cell types toward 2019-nCoV infection. We leveraged the recently published high-throughput single-cell expression study to evaluate cell type-specific expression levels of the well-known viral entry host genes. Our meta-analysis revealed that a subset of SUSs that are enriched for cytoskeleton regulatory proteins are the most vulnerable cell type to the 2019-nCoV infection, followed by a minor population of BGCs and olfactory stem cells (OSCs, GBCs and HBCs). Aside from humans, we also pinpointed four at-risk mammalian species with high susceptibility to 2019-nCoV infection and the potential of experiencing an infection-mediated loss of olfaction.
Figure 1.
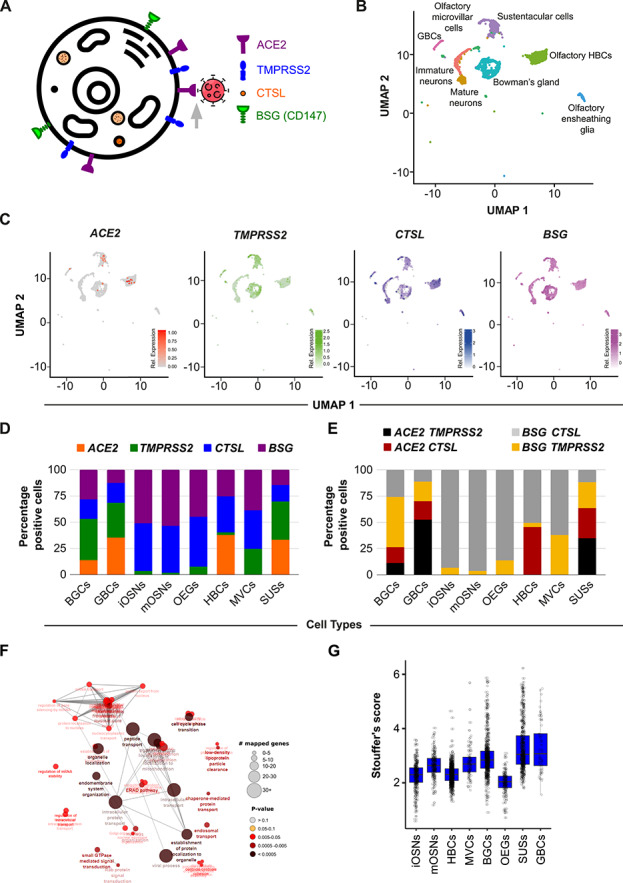
OSNs do not express 2019-nCoV entry genes. (A) Schematic diagram depicting the subcellular localization of the known 2019-nCoV entry host proteins. (B) UMAP-based embedding of single-cell expression profiles represents the distinct cell types of the olfactory epithelium (C) UMAP-based embedding portrays the relative expression of indicated transcripts in the distinct cell types of the human olfactory epithelium. (D) Stacked bar graphs representing the relative proportions of cells (percent normalized) expressing the indicated 2019-nCoV entry-associated genes. (E) Stacked bar graph representing the relative proportion of cells (percent normalized) co-expressing the known host receptor (ACE2 or BSG) and cellular protease (TMPRSS2 or CTSL). (F) Functional enrichment analysis of virus–human protein–protein interactome genes reliably identified in olfactory epithelial cell types. (G) Box plot depicting Stouffer’s score computed based on virus–human protein–protein interaction-related genes across the indicated cell types of the olfactory epithelium.
Materials and methods
Single-cell RNA-sequencing analysis
The raw read counts of the single-cell RNA-sequencing datasets were downloaded from GEO (GSE139522) [34]. We performed the majority of our analyses including cell/gene filtering, clustering and differential expression analysis, using the widely used Seurat software suite [35]. Inbuilt functions NormalizeData(), FindVariableFeatures(), ScaleData(), RunPCA(), DimPlot(), FindNeighbors() and FindClusters() were used for the various standard steps of single-cell expression data analysis. Each cluster could be unequivocally mapped to a known cell type, based on the markers reported in the original study (Figure S1A and B) [34]. Differential gene expression was performed using the Poisson method, an inbuilt function of the Seurat software suite. To ensure reproducibility, we constructed average expression vectors for different predefined categories of cells (all eight olfactory cell types and ACE2-positive cells/TMPRSS2-positive cells/CTSL-positive cells/BSG (CD147)-positive cells separately) across two biological replicates, as available from the concerned study [34]. The extent of the linear relationship between each of the normalized expression vector pairs was computed using Pearson’s correlation coefficient. We also performed a chi-square test for comparing the relative proportions of various 2019-nCoV-susceptible cell subpopulations between the biological replicates.
Estimating the extent of host–virus protein–protein interactions across cell types
The host–virus protein–protein interactions were obtained from a recent study [36]. For cell and gene filtering, we used the FilterCells() and FilterGenes() functions, respectively, from the dropClust pipeline, with the default parameter values [37, 38]. The reduced expression matrix thus obtained was subjected to median normalization. We log2-transformed the normalized expression estimates after adding 1 as pseudo-count. Next, we retained those N genes, for which the corresponding proteins existed among the host proteins reported by the study. The second pass of cell filtering was performed to retain cells that expressed at least 10% of these genes. For each gene, the log-normalized expression estimates were converted into Z-scores. For each cell, a combined Stouffer’s Z-score 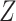 was computed as
was computed as 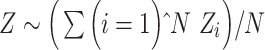 , where
, where 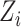 denotes the cell-specific Z-score corresponding to
denotes the cell-specific Z-score corresponding to 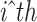 gene and
gene and 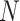 denotes the number of genes common between the expression and the protein–protein interaction data. A Stouffer’s score, in this context, reflects the extents of protein–protein interaction in a cell. One-sided Wilcoxon rank-sum test was performed to assess the statistical significance of cell type-specific putative enrichment of host–virus protein–protein interactions (Figure S1E).
denotes the number of genes common between the expression and the protein–protein interaction data. A Stouffer’s score, in this context, reflects the extents of protein–protein interaction in a cell. One-sided Wilcoxon rank-sum test was performed to assess the statistical significance of cell type-specific putative enrichment of host–virus protein–protein interactions (Figure S1E).
Analysis of bulk RNA-sequencing dataset
Uniformly processed bulk RNA-sequencing data containing transcriptomic profiles of whole olfactory mucosa from five mammalian species, i.e. human, monkey, marmoset, mouse and rat, were obtained from a recent publication from Saraiva and colleagues [39]. Log-transformed FPKM values were used for plotting the bar charts. The student’s t-test was used to calculate the differences in the mean values across the species. A P-value <0.05/<0.01/<0.001/<0.0001 is denoted as */**/***/****.
Construction of the phylogenetic tree
To construct the phylogenetic tree, the protein sequences of ACE2 of five mammalian species were downloaded from the NCBI database. The phylogenetic tree was constructed using an online web server (http://www.phylogeny.fr/) [40]. Protein sequences were supplied to the web server in FASTA format. Multiple sequence alignment was performed using MUSCLE (version 3.8.31). For refining the alignments, Gblocks (version 0.91b) was used with a minimum block length of 10, and no allowed gap positions were used. Whelan and Goldman amino acid substitution model was used (available in PhyML 3.1/3.0 aLRT) [41], where the number of substitution rate categories was set to 4. Finally, the tree was rendered using TreeDyn (version 198.3) [42].
Homology modeling and molecular docking
The protein sequences of ACE2 receptors of all species were obtained from the NCBI database (rat, XP_032746145.1; mouse, BAB40431.1; marmoset, XP_008987241.1; macaque, NP_001129168.1). For homology modeling, the human ACE2 was used as the template (PDB ID: 6VW1) [43]. The 3D structures were generated by using Modeller v9.24 [44]. The quality of the models was assessed by the Discrete Optimised Protein Energy (DOPE) score, provided by the Modeller as well as by the Ramachandran plots, produced by RAMPAGE. After obtaining the refined models, we next performed docking experiments involving protein–protein interactions between the spike RBD of 2019-nCoV and the host-specific ACE2 receptors. We used the HADDOCK 2.4 web server for molecular docking experiments [45]. The previously known residues involved in the interaction were used to define the active and passive residues of the proteins. HADDOCK produces ranked clusters of protein–protein complexes based on the HADDOCK score (1.0 Evdw + 0.2 Eelec + 1.0 Edesol + 0.1 EAIR). Notably, the HADDOCK score consists of a combination of empirical (desolvation, buried surface area) and energy (electrostatic, van der Waals) terms. The top resultant complexes were then processed by PROtein binDIng enerGY prediction (PRODIGY) web platform to evaluate their binding energy [46]. As a control, docking of human ACE2 was performed with RBD of SARS-CoV (PDB ID: 2AJF) while using the same docking parameters. Pairwise Mann–Whitney test was performed to compute the statistical significance.
Multiple sequence alignment
The sequence alignment of ACE2 proteins across five different mammalian species (human, rat, mouse, macaque, marmoset) enabled the identification of highly conserved residues (i.e. same residue at the same position in all five species) and partially conserved residues (i.e. replaced by an amino acid with similar biochemical properties or, in other words, a conservative or semiconservative replacement). Multiple sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) [47].
Code availability
Source code used for the single-cell expression data analysis is accessible from the GitHub repository (https://github.com/krishan57gupta/The-cellular-basis-of-the-loss-of-smell-in-2019-nCoV-infected-individuals).
Results
Divergent expression dynamics of viral entry genes across olfactory cell subpopulations
We evaluated the expression of a panel of the known viral entry transcripts (ACE2, TMPRSS2, BSG/CD147 and CTSL) in 3906 olfactory epithelium originated single cells from the recent report by Durante and colleagues [34], collectively entailing eight distinct olfactory cell types, namely, HBCs, MVCs, BGCs, GBCs, OEGs, SUSs, iOSNs and mOSNs. We performed unsupervised clustering of the individual cells using the Seurat software suite [35]. The clusters thus obtained were unambiguously mapped to specific cell types based on previously known markers (Figure 1B and Figure S1A, B) [34]. All the four well-known viral entry transcripts showed distinct expression patterns across each of the eight cell types. In the case of ACE2, we observed sparse expression levels, primarily restricted to four cell types, i.e. SUSs, BGCs, GBCs and HBCs. Notably, these four cell types collectively constitute less than 1% (32 out of 3906) of the total analyzed cell population. Conversely, transcripts from TMPRSS2, BSG and CTSL were observed at relatively higher concentrations across all the cell types (Figure 1C). Besides host receptor binding, efficient entry of 2019-nCoV also requires priming of the viral S protein via host proteases [16]. As such, we next investigated the cellular co-occurrence of these essential moieties. Our co-expression analysis at the single-cell resolution further revealed the higher infection susceptibility in a subset of SUSs across all combinations (Figure 1D and E). Notably, due to the lack of direct evidence for the BSG-mediated viral entry into the host cell [15], for further analysis, we focused on the expression dynamics of ACE2 and TMPRSS2. Next, we characterized the phenotypic divergence between ACE2+; TMPRSS2+ and ACE2−; TMPRSS2− subpopulations of the SUSs by examining the differentially expressed genes (DEGs) (Figure S1C). Functional enrichment analysis of the significant DEGs (log2FC ≥ 1 or ≤ −1; FDR < 0.05) revealed the enrichment of the cytoskeleton regulation genes in ACE2+; TMPRSS2+ double-positive SUSs (Figure S1D). Notably, certain replication machinery components of SARS-CoV, a virus similar in properties to that of 2019-nCoV, utilize microtubule-associated intracellular transport [48].
We used an orthogonal approach leveraging host–virus protein interactome, to identify the most vulnerable olfactory cell types for 2019-nCoV infection. For this, we developed a novel strategy to overlay host–virus protein interactome on cell type-specific expression signatures (section Materials and methods). Based on the interactome enrichment analysis, we ranked the various olfactory cell types. In line with our previous analyses, the SUSs were found to be maximally susceptible to viral infection (Figure 1F and G; Figure S1E). Importantly, to ensure the reproducibility of our findings, we performed the reproducibility analysis on all the single olfactory cell types obtained from two biological replicates (patients 2 and 3 of Durante et al. 2020 [34]). For each subpopulation, our analysis revealed highly reproducible expression patterns across the replicates (Figure S2). Moreover, similar results were obtained when the relative proportions of the 2019-nCoV-susceptible cells were compared between the biological replicates (ACE2 and TMPRSS2 double-positive cells; chi-square value = 2.75, P-value = 0.43). In summary, while the OSNs largely lack the host-specific proteins essential for the cellular entry of 2019-nCoV, the supporting and the stem cell subpopulations of the olfactory epithelium are potentially highly susceptible to the viral infection.
Comparable expression levels and binding affinity of ACE2 toward viral spike protein across five mammalian species
The rate of transmission of 2019-nCoV is remarkably higher as compared to the related SARS-CoV [49]. Although it has been speculated that the 2019-nCoV is transmitted to humans from animal sources [50], little is known about the capability of the other mammal species to act as carriers. Notably, in a recent report, monkeys have been confirmed as potential carriers of 2019-nCoV [51]. We studied if other mammalian species are also at the risk of developing 2019-nCoV-mediated loss of olfaction. To test this, we first estimated the messenger RNA levels of ACE2 and TMPRSS2 transcripts in the bulk RNA-Seq profiles of the whole olfactory mucosa of five mammalian species. Our results suggest a comparable expression of these two viral entry genes (ACE2 and TMPRSS2) among all the species (Figure 2A and B). Next, in order to gain direct evidence of the molecular interactions between the viral S protein and ACE2, we first modeled and refined the three-dimensional stable protein structure of ACE2 homologs from all the four mammalian species (Figure S3A-D). To achieve this, we used the recently solved human ACE2 protein structure as a template (Figure S3A-D). Next, we performed molecular docking between the viral RBD [43] and the modeled ACE2 proteins specific to the individual species (Figure 2C and D). A comparison of the binding parameters further revealed similar binding affinities across all tested pairs, suggesting similar vulnerability for 2019-nCoV infection in these species (Figure 2E-G). Importantly, to ensure the robustness of the docking experiments, we compared the HADDOCK scores obtained from docking of human ACE2 with RBD of SARS-CoV and 2019-nCoV. Notably, a recent report experimentally estimated the EC50 (half-maximal effective concentration) values of the aforementioned interactions (human ACE2 and viral RBD domains) [52] and observed preferable binding of 2019-nCoV with that of human ACE2, as compared to SARS-CoV (Figure S3E). We also observed significantly lower HADDOCK scores in the docking simulations of human ACE2 and 2019-nCoV (pairwise Mann–Whitney U-test, P-value <0.0001), which is in line with the published experimental results [52]. Collectively, all these analyses suggest that similar to humans, the olfactory system of other mammals could also be at potential risk of 2019-nCoV infection.
Figure 2.
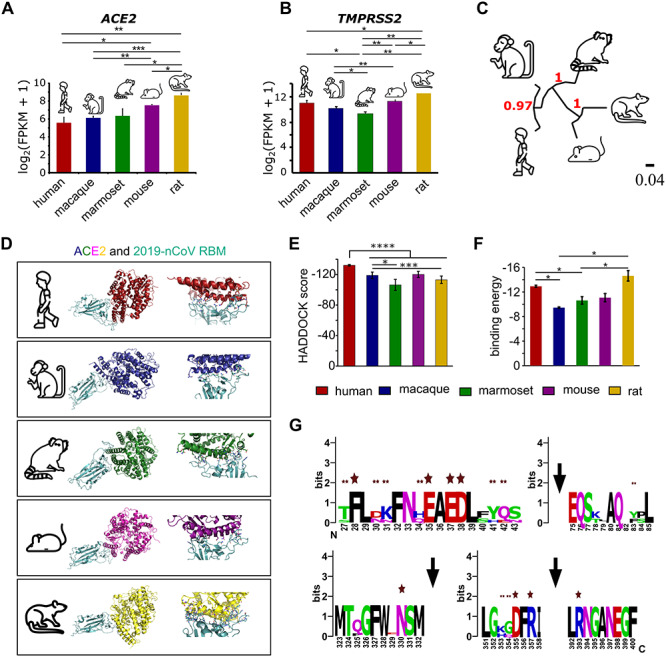
Multifactor analysis involving gene-expression and molecular docking highlights the potential risk of olfactory dysfunction in other mammals. (A) Bar graph depicting the relative abundance of ACE2 in the bulk RNA sequencing of the whole olfactory mucosa of five indicated mammalian species. Bars represent the mean values, the error bars represent the standard deviation, and asterisks represent statistical significance. (B) Bar graph depicting the relative abundance of TMPRSS2 in the bulk RNA sequencing of the whole olfactory mucosa of five indicated mammalian species. The bar represents the mean values, the error bars represent the standard deviation, and asterisks represent statistical significance. (C) Phylogenetic tree depicting the ACE2 sequence similarities between five mammalian species. (D) Protein structures depicting the molecular interactions between ACE2 proteins and the RBD domain of 2019-nCoV estimated using computationally assisted molecular docking. Structure of 2019-nCoV RBD (pale cyan) complexed with its receptor ACE2 (distinct color for different species). (E) Bar graph depicting the HADDOCK scores under the indicated conditions. Error bars represent the standard deviation of the estimates, and asterisks represent statistical significance. (F) Bar graph representing the binding energies of the interaction between the 2019-nCoV receptor-binding domain and ACE2 receptor in the indicated species. Error bars represent the standard deviation of the estimates, and asterisks represent statistical significance. (G) Web logo representing the key conserved amino acids of ACE2 of five mammalian species. Single and double asterisks represent highly and partially conserved known interacting residues, respectively.
Discussion and future directions
In addition to the infection-induced multi-organ dysfunction, a major bottleneck in combating the pandemic outbreak of 2019-nCoV is the availability and accessibility of the diagnostic methods to masses worldwide. Although major initiatives have been taken to develop 2019-nCoV centric molecular diagnostic kits, their fabrication, manufacturing, mass dissemination and adoption are likely to be time consuming. Recently, multiple clinical studies have reported the abrupt loss of smell and taste in a large number of 2019-nCoV-infected individuals [23–25], thereby, collectively, reinforcing its potential application as the first line of diagnostics in the patients exhibiting other 2019-nCoV-related hallmark symptoms. In pursuit of this, the Global Consortium of Chemosensory Researchers (GCCR) has initiated a worldwide scientific study to assess the possible relationships between respiratory illness and its effects on smell and taste. Similarly, other studies also point toward the sudden loss of olfaction as the first manifestation in the confirmed 2019-nCoV-infected patients [23–25]. Additionally, the symptom survey of 2019-nCoV-infected patients also revealed the loss of smell as a stronger predictor of positive diagnosis than the self-reported fever [23]. Our study aims to underscore the potential cellular basis of the loss of olfaction, by examining the olfactory epithelium-specific cell types based on the expression levels of the host-specific viral entry moieties as well as the burden of host–virus protein–protein interactions. Our analyses suggest that the loss of smell in the infected patients might not be due to the direct impairment of the OSNs. Instead, SUSs, BGCs and OSCs (HBCs and GBCs) exhibit the molecular makeup that makes the cells susceptible to viral infection (Figure 3). This conclusion was drawn based on a consensus approach involving gene expression as well as host–pathogen protein interactome. Importantly, since all our findings are largely substantiated by in silico analysis, one cannot obviate the limitations of the single-cell transcriptomics assay such as sampling bias or high dropout rates [53]. While the identification of the 2019-nCoV infection-susceptible cell types of the olfactory epithelium is characterized based on a handful of known viral entry moieties of the host cells, one cannot rule out the possible involvement of currently uncharacterized host cell surface receptors or proteases which may facilitate the viral entry into the host cells.
Figure 3.
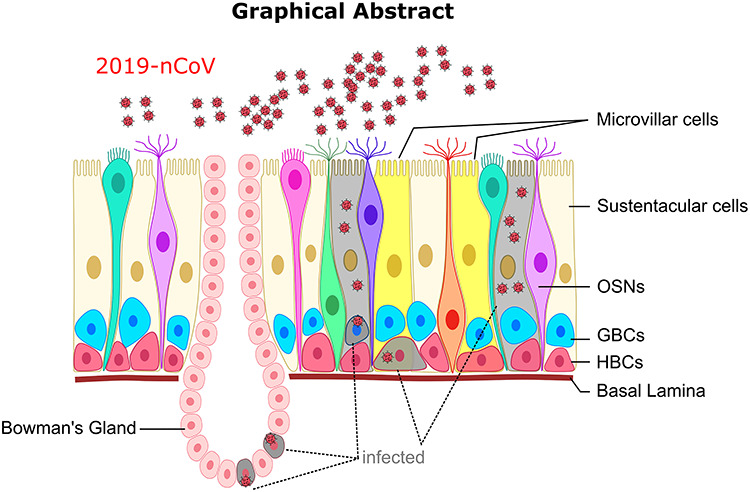
Graphical representation of the key findings.
The direct sensory roles of the SUSs are largely elusive; however, they are known to provide metabolic and physical support to the olfactory epithelium [54]. Particularly the SUSs are known to be involved in secretion [55], endocytosis [56] and cytochrome P-450-mediated detoxification [57]. Moreover, as glia-like cells, they impart critical functionality related to phagocytosis of dead cells [58] and regulation of the ionic exchange with the extracellular regions [59, 60]. Hegg et al. [61] identified a key function of SUSs in establishing communication between neurons, basal cells and SUSs themselves. The authors identified that the activation of G-protein-coupled receptors, particularly the P2Y purinergic receptor and the muscarinic acetylcholine receptor, induces calcium oscillations in SUSs. They further provided mechanistic insights by using pharmacological interventions and showed the involvement of phospholipase C (PLC) pathways in triggering the calcium increase. Moreover, studies utilizing Xenopus laevis as a model organism revealed the implication of intraepithelial purinergic signaling, possibly via SUS, in the proliferation of the olfactory stem cells, thereby influencing odorant detection [62]. Interestingly, SUSs are also known to mediate intraepithelial signaling and could potentially modulate the odor detection thresholds of OSNs using endocannabinoids. Czesnik et al. in 2007 used CB1 receptor-specific antagonists on Xenopus larvae and observed modulation in the odor-evoked calcium changes within the mOSNs [63]. Moreover, in a subsequent study, Breunig et al. revealed that 2-arachidonoylglycerol (2-AG), an endocannabinoid, is synthesized by the glia-like SUSs, and its synthesis largely depends on the hunger state of the organism [64]. Interestingly, the authors have also observed decreased detection thresholds to food odors in the OSNs in the presence of 2-AG, suggesting a plausible mechanism by which SUS can indirectly influence the receptor neurons [64]. Another putative mechanism that could induce such a starking phenotype is the infection in the blood vessels (endothelial cells), which may also indirectly influence the odor detection thresholds [21].
Lastly, although SUSs are not directly involved in odor detection, one cannot rule out their plausible involvement in mediating the loss of smell symptom in 2019-nCoV-infected patients, as they are the key maintainers of the olfactory organ homeostasis and its architecture [30, 65–67]. These assumptions have been substantiated by rigorous experiments on rodents, where damage to SUSs and BGCs induces architectural damage in the olfactory epithelium [54, 60].
Notably, in addition to SUSs, our results also highlight the viral infection susceptibility in minor subpopulations of BGCs and OSCs. BGCs play a vital role in maintaining the optimal functionality of the olfactory system. First, they produce a number of olfactory binding proteins to facilitate the transport of odorants to olfactory receptor cells. Second, they also secrete mucus which protects the olfactory epithelium from drying out and therefore indirectly assists odor recognition by mOSNs [54]. Similarly, while OSCs are not known to have any direct role in odorant detection, they play a crucial role in the regeneration of the olfactory epithelium upon lesions [32, 68]. Notably, injury models involving the direct loss of SUSs have been shown to activate the HBCs which in turn proliferate and replenish the lost cells, thereby reconstituting the olfactory epithelium homeostasis [68]. Mechanistically, loss of Notch signaling pathways between SUSs and HBCs leads to the breakdown of mitotic dormancy of HBCs by downregulating tumor protein p63 [69]. In light of these critical functional roles, we speculate that the apparent loss of smell could be a result of viral load in SUSs, BGCs and OSCs. Due to their apical localization in the olfactory epithelium, 2019-nCoV may first infect SUS cells, leading to the partial/complete breakdown of the olfactory architecture, resulting in the decline in olfaction. Moreover, the intensity of this phenotype further exaggerates due to a failure in the repair response because of the subsequent infection in the OSCs. In addition to this, our analysis on four additional mammalian species also suggests that the 2019-nCoV-mediated loss of smell phenotypes is not restricted to humans but may also impact other mammalian species. Collectively, our study provides the first line of evidence that a subpopulation of olfactory cells is potentially equipped with host-specific viral entry moieties, which can be exploited by the virus for its entry.
Key Points
Loss of smell (anosmia) is emerging as one of the key early symptoms in 2019-nCoV-infected individuals worldwide, indicating an atypical direct impact on the olfactory organ.
As olfactory receptor neurons are not directly targeted by the 2019-nCoV, a direct explanation of the loss of smell phenotype is not so obvious.
2019-nCoV-human protein–protein interaction-based bioinformatics analysis revealed the highest infection susceptibility in the SUSs.
2019-nCoV infection-mediated compromise in abundance or cellular function of SUSs (supporting cells), olfactory stem cells (OSCs) and Bowman’s gland cells of the olfactory epithelium are the most probable causes of anosmia.
A multifactor analysis involving the gene-expression analysis and molecular docking collectively indicates the potential risk of olfactory dysfunction in four additional mammalian species.
Supplementary Material
Acknowledgements
We would like to thank the IT-HelpDesk team of IIIT-Delhi for providing assistance with the computational resources. We thank all the members of the Ahuja lab for their intellectual contributions at various stages of this project.
Krishan Gupta is a PhD student of Indraprastha Institute of Information Technology, Delhi. His research interests include developing methods for the single-cell RNA-sequencing data analysis.
Sanjay Kumar Mohanty is a student of Master's in Bioinformatics, presently working on his thesis dissertation at Indraprastha Institute of Information Technology, Delhi.
Aayushi Mittal is a PhD student at the Indraprastha Institute of Information Technology, Delhi. Her research interests include gene-expression regulation and epigenetics.
Siddhant Kalra is an M-Tech student (Master of Technology in Computational Biology) at the Indraprastha Institute of Information Technology, Delhi. His research interests include olfaction and computational neuroscience.
Suvendu Kumar is a student of Master's in Bioinformatics, presently working on his thesis dissertation at the Indraprastha Institute of Information Technology, Delhi.
Tripti Mishra PhD is a data scientist at the Pathfinder Research and Training Foundation.
Jatin Ahuja DNB, DM is a clinician with superspecialization (Doctorate of Medicine) in Infectious diseases.
Debarka Sengupta PhD is an Assistant Professor at the Department of Computational Biology, and Department of Computer Science at the Indraprastha Institute of Information Technology. His research interests include single-cell genomics, liquid biopsy and machine learning.
Gaurav Ahuja PhD is an assistant professor at the Department of Computational Biology at the Indraprastha Institute of Information Technology-Delhi (IIIT-Delhi). His research interests include decoding the gene regulatory mechanisms mediating diseases onset/progression, cell-fate transitions and cellular response to ligands.
Contributor Information
Krishan Gupta, Indraprastha Institute of Information Technology, Delhi.
Sanjay Kumar Mohanty, Indraprastha Institute of Information Technology, Delhi.
Aayushi Mittal, Indraprastha Institute of Information Technology, Delhi.
Siddhant Kalra, Indraprastha Institute of Information Technology, Delhi.
Suvendu Kumar, Indraprastha Institute of Information Technology, Delhi.
Tripti Mishra, Indraprastha Institute of Information Technology, Delhi.
Jatin Ahuja, Indraprastha Institute of Information Technology, Delhi.
Debarka Sengupta, Indraprastha Institute of Information Technology, Delhi.
Gaurav Ahuja, Indraprastha Institute of Information Technology, Delhi.
Author contributions
The study was conceived by G.A. and D.S. Experimental workflows were designed by G.A., D.S. and T.M. and performed by K.G., S.K.M., S.K., A.M., and S. Clinical insights were provided by J.A. Illustrations were drafted by G.A. and A.M. G.A. and D.S. wrote the paper. All authors have read and approved the manuscript.
Funding
The Ahuja lab is supported by the Intramural Startup grant from Indraprastha Institute of Information Technology-Delhi (IIIT-Delhi) and Ramalingaswami Re-entry Fellowship, a re-entry scheme of the Department of Biotechnology (DBT), Ministry of Science and Technology, Govt. of India. The Sengupta lab is funded by the INSPIRE faculty grant from the Department of Science and Technology (DST), India.
References
- 1. Khot WY, Nadkar MY. The 2019 novel coronavirus outbreak - a global threat. J Assoc Physicians India 2020;68:67–71. [PubMed] [Google Scholar]
- 2. Cascella M, Rajnik M, Cuomo A, et al. Features, evaluation and treatment coronavirus (COVID-19). In: StatPearls 2020. Treasure Island, FL: StatPearls Publishing. [PubMed] [Google Scholar]
- 3. Lewis D. Coronavirus outbreak: what’s next? Nature 2020;578:15–6. [DOI] [PubMed] [Google Scholar]
- 4. Fadel M, Salomon J, Descatha A. Coronavirus outbreak: the role of companies in preparedness and responses. Lancet Planet Health 2020;5:e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paraskevis D, Kostaki EG, Magiorkinis G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol 2020;79:104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 2020;92:418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan JF-W, Kok K-H, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 2020;9:221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hofmann H, Pyrc K, Hoek L, et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 2005;102:7988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020;368:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hasan A, Paray BA, Hussain A, et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J Biomol Struct Dyn 2020;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang K, Chen W, Zhou Y-S, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. doi: 10.1101/2020.03.14.988345. [DOI] [Google Scholar]
- 16. Padmanabhan P, Desikan R, Dixit N. Targeting TMPRSS2 and Cathepsin B/L Together May Be Synergistic Against SARS-CoV-2 Infection, 2020. ChemRxiv Preprint. doi: 10.26434/chemrxiv.12213125.v2. [DOI] [PMC free article] [PubMed]
- 17. Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y, Zhao Z, Wang Y, et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv 2020. doi: 10.1101/2020.01.26.919985. [DOI] [Google Scholar]
- 19. Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019-nCoV infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv 2020. doi: 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- 20. Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in Cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv 2020. doi: 10.1101/2020.02.03.931766. [DOI] [Google Scholar]
- 21. Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020;395:1417–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaim S, Chong JH, Sankaranarayanan V, et al. COVID-19 and multiorgan response. Curr Probl Cardiol 2020;45:100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menni C, Valdes A, Freydin MB, et al. Loss of smell and taste in combination with other symptoms is a strong predictor of COVID-19 infection medRxiv, 2020. doi: 10.1101/2020.04.05.20048421. [DOI]
- 24. Yan CH, Faraji F, Prajapati DP, et al. Self-reported olfactory loss associates with outpatient clinical course in COVID-19. Int Forum Allergy Rhinol 2020;10:821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jerome L, Pierre C, Carlos C-E, et al. Objective olfactory testing in patients presenting with sudden onset olfactory dysfunction as the first manifestation of confirmed COVID-19 infection. medRxiv 2020. doi: 10.1101/2020.04.15.20066472. [DOI] [Google Scholar]
- 26. Chen CR, Kachramanoglou C, Li D, et al. Anatomy and cellular constituents of the human olfactory mucosa: a review. J Neurol Surg B Skull Base 2014;75:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahuja G, Bozorg Nia S, Zapilko V, et al. Kappe neurons, a novel population of olfactory sensory neurons. Sci Rep 2014;4:4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci U S A 2004;101:2584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang F. Sustentacular cell enwrapment of olfactory receptor neuronal dendrites: an update. Genes (Basel) 2020;11:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moran DT, Rowley JC, Jafek BW, et al. The fine structure of the olfactory mucosa in man. J Neurocytol 1982;11:721–46. [DOI] [PubMed] [Google Scholar]
- 31. Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec 2002;269:33–49. [DOI] [PubMed] [Google Scholar]
- 32. Joiner AM, Green WW, McIntyre JC, et al. Primary cilia on horizontal basal cells regulate regeneration of the olfactory epithelium. J Neurosci 2015;35:13761–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwob JE, Jang W, Holbrook EH, et al. Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol 2017;525:1034–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durante MA, Kurtenbach S, Sargi ZB, et al. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat Neurosci 2020;23:323–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sinha D, Sinha P, Saha R, et al. Improved dropClust R package with integrative analysis support for scRNA-seq data. Bioinformatics 2019;36:1946–47. [DOI] [PubMed] [Google Scholar]
- 38. Sinha D, Kumar A, Kumar H, et al. dropClust: efficient clustering of ultra-large scRNA-seq data. Nucleic Acids Res 2018;46:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saraiva LR, Riveros-McKay F, Mezzavilla M, et al. A transcriptomic atlas of mammalian olfactory mucosae reveals an evolutionary influence on food odor detection in humans. Sci Adv 2019;5:eaax0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36:W465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 2001;18:691–9. [DOI] [PubMed] [Google Scholar]
- 42. Chevenet F, Brun C, Bañuls A-L, et al. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 2006;7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020;581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993;234:779–815. [DOI] [PubMed] [Google Scholar]
- 45. Zundert GCP, Rodrigues JPGLM, Trellet M, et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 2016;428:720–5. [DOI] [PubMed] [Google Scholar]
- 46. Xue LC, Rodrigues JP, Kastritis PL, et al. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016;32:3676–8. [DOI] [PubMed] [Google Scholar]
- 47. Sievers F, Higgins DG. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 2014;1079:105–16. [DOI] [PubMed] [Google Scholar]
- 48. Naghavi MH, Walsh D. Microtubule regulation and function during virus infection. J Virol 2017;91:e00538-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 2020;382:1564–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong MC, Javornik Cregeen SJ, Ajami NJ, et al. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv 2020. doi: 10.1101/2020.02.07.939207. [DOI] [Google Scholar]
- 51. Infection with Novel Coronavirus (SARS-CoV-2) Causes Pneumonia in the Rhesus Macaques, 2020;1–8. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed]
- 52. Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020;17:613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen G, Ning B, Shi T. Single-cell RNA-Seq technologies and related computational data analysis. Front Genet 2019;10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Getchell ML, Getchell TV. Fine structural aspects of secretion and extrinsic innervation in the olfactory mucosa. Microsc Res Tech 1992;23:111–27. [DOI] [PubMed] [Google Scholar]
- 55. Doty RL. Introduction and Historical Perspective. In: Handbook of Olfaction and Gustation, 2015;1–36.
- 56. Bannister LH, Dodson HC. Endocytic pathways in the olfactory and vomeronasal epithelia of the mouse: ultrastructure and uptake of tracers. Microsc Res Tech 1992;23:128–41. [DOI] [PubMed] [Google Scholar]
- 57. Dahl AR, Hadley WM, Hahn FF, et al. Cytochrome P-450-dependent monooxygenases in olfactory epithelium of dogs: possible role in tumorigenicity. Science 1982;216:57–9. [DOI] [PubMed] [Google Scholar]
- 58. Suzuki Y, Takeda M, Farbman AI. Supporting cells as phagocytes in the olfactory epithelium after bulbectomy. J Comp Neurol 1996;376:509–17. [DOI] [PubMed] [Google Scholar]
- 59. Breipohl W, Laugwitz HJ, Bornfeld N. Topological relations between the dendrites of olfactory sensory cells and sustentacular cells in different vertebrates. An ultrastructural study. J Anat 1974;117:89–94. [PMC free article] [PubMed] [Google Scholar]
- 60. Rafols JA, Getchell TV. Morphological relations between the receptor neurons, sustentacular cells and Schwann cells in the olfactory mucosa of the salamander. Anat Rec 1983;206:87–101. [DOI] [PubMed] [Google Scholar]
- 61. Hegg CC, Irwin M, Lucero MT. Calcium store-mediated signaling in sustentacular cells of the mouse olfactory epithelium. Glia 2009;57:634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hassenklöver T, Schwartz P, Schild D, et al. Purinergic signaling regulates cell proliferation of olfactory epithelium progenitors. Stem Cells 2009;27:2022–31. [DOI] [PubMed] [Google Scholar]
- 63. Czesnik D, Schild D, Kuduz J, et al. Cannabinoid action in the olfactory epithelium. Proc Natl Acad Sci U S A 2007;104:2967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Breunig E, Manzini I, Piscitelli F, et al. The endocannabinoid 2-arachidonoyl-glycerol controls odor sensitivity in larvae of Xenopus laevis. J Neurosci 2010;30:8965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huard JM, Youngentob SL, Goldstein BJ, et al. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol 1998;400:469–86. [PubMed] [Google Scholar]
- 66. Carr VM, Farbman AI, Colletti LM, et al. Identification of a new non-neuronal cell type in rat olfactory epithelium. Neuroscience 1991;45:433–49. [DOI] [PubMed] [Google Scholar]
- 67. Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 2007;10:720–6. [DOI] [PubMed] [Google Scholar]
- 68. Iwai N, Zhou Z, Roop DR, et al. Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells 2008;26:1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Herrick DB, Lin B, Peterson J, et al. Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci U S A 2017;114:E5589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


