Abstract
Accurate understanding of the global spread of emerging viruses is critically important for public health response and for anticipating and preventing future outbreaks. Here, we elucidate when, where and how the earliest sustained SARS-CoV-2 transmission networks became established in Europe and the United States (US). Our results refute prior findings erroneously linking cases in January 2020 with outbreaks that occurred weeks later. Instead, rapid interventions successfully prevented onward transmission of those early cases in Germany and Washington State. Other, later introductions of the virus from China to both Italy and Washington State founded the earliest sustained European and US transmission networks. Our analyses reveal an extended period of missed opportunity when intensive testing and contact tracing could have prevented SARS-CoV-2 from becoming established in the US and Europe.
One sentence summary:
Sustained SARS-CoV-2 transmission networks became established in Europe and the US several weeks later than previously estimated.
Introduction
The emergence in late 2019 of SARS-CoV-2, the agent of COVID-19, has ignited a global health crisis unparalleled since the 1918 influenza pandemic. Rapid sharing of viral sequence data that began within weeks of identification of the virus (1) allowed the development of diagnostic tests crucial to control efforts and initiated research enabling candidate vaccines and therapies. These genomes also precipitated a worldwide effort of viral genomic sequencing and analyses unprecedented in scale and pace. At time of writing (May 15th, 2020) there are already 25,181 complete genomes available.
Embedded within this assemblage of genomic data is crucial information about the nature and history of the pandemic. Here, we investigate fundamental questions about when, where and how this new virus established itself globally—scientific questions that have become inextricably linked to important societal and policy concerns. The combination of the relatively slow rate of SARS-CoV-2 evolution, its rapid dissemination within and between locations, and extremely unrepresentative sampling means that naive interpretation of phylogenetic analyses can lead to serious error. However, we show that with the integration of multiple sources of information, careful consideration of evolutionary process and pattern, and cutting-edge technology, key events in the unfolding of the pandemic, which have been the subject of conjecture and controversy but little quantitative study, can be resolved.
Here we consider two of the earliest known introductions—successful or not—of SARS-CoV-2 into both Europe and the US.
The first patient to be diagnosed with COVID-19 in the US, designated ‘WA1’, was a Chinese national who travelled from Wuhan, in Hubei Province, China, to Sea-Tac International Airport near Seattle, Washington State, arriving on January 15th, 2020 (2). This individual was attuned to Centers for Disease Control and Prevention (CDC) messaging about the new pneumonial disease circulating in Wuhan and promptly sought medical care upon becoming symptomatic with COVID-19, receiving a sequence-confirmed diagnosis and becoming the first US patient to have a SARS-CoV-2 genome sequenced, sampled on January 19th, 2020 (3). Efficient contact tracing measures were enacted by local health authorities and the patient and public health authorities worked together in an exemplary fashion to limit opportunities for spread (3). No onward transmission was detected after exhaustive follow-up in what appeared to be successful containment of the first known incursion of the virus in the US (3).
On February 29th, 2020, a SARS-CoV-2 genome was reported (4) from a second Washington State patient, ‘WA2’, whose virus had been sampled on February 24th as part of a community surveillance study of respiratory viruses (5). The report’s authors calculated a high probability that WA2 was a direct descendent of WA1, coming to the surprising conclusion that there had by that point already been six weeks of cryptic circulation of the virus in Washington State (4). The finding, described in a lengthy Twitter thread on February 29th, fundamentally altered the picture of the SARS-CoV-2 situation in the US, and seemed to show how the power of genomic epidemiology could be harnessed to uncover hidden epidemic dynamics and inform policy making in real time (6, 7). The COVID-19 pandemic represents the first major global disease event to emerge in the age of social media, and the urgent need for timely information to inform public health decisions has frequently outpaced standard peer-review processes. As a result, the findings played a decisive role in Washington State’s early adoption of intensive social distancing efforts, which, in turn, appeared to explain Washington State’s relative success in controlling the outbreak, compared with states that delayed, such as New York (8).
In Europe, an employee of the auto supplier Webasto visited the company’s headquarters in Bavaria, Germany, from Shanghai, China, on January 20th, 2020 (9). She had been infected with SARS-CoV-2 in Shanghai (after her parents had visited from Wuhan) (10) and transmitted the virus to a German man who tested positive on January 27th (11) and whose viral genome (‘BavPat1’) was sampled on January 28th (10). All told, the outbreak infected 16 employees but was apparently contained through rapid testing and isolation, in efforts that were lauded as an impressive, early display of how to stop the virus (9).
Weeks later, however, Italy’s first major outbreak in Lombardy was associated with viruses closely related to BavPat1, differing by just one nucleotide in the nearly 30,000 nucleotide genome. At a time when social and news media outpaced scientific peer-review, a narrative took hold that the virus from Germany had in fact not been contained but had been transmitting undetected for weeks and had been carried to Italy by an infected German (9, 12). In addition to igniting a devastating outbreak in Italy, this particular lineage (B.1) subsequently spread widely across Europe, seeding outbreaks in many countries (13).
Importantly, this lineage also spread to North America, establishing an early beachhead in New York City (14, 15). Furthermore, the B.1 lineage harbors a D614G mutation in the Spike protein that has been controversially claimed to make the virus more contagious (16). Greater clarity about the role of that initial outbreak in Germany in the emergence of SARS-CoV-2 in Italy—and then the rest of Europe and the US—has critical implications for individuals involved in the German outbreak and response as well as for the broader feasibility of controlling the virus through gold standard public health responses.
Results
Emergence of SARS-CoV-2 in Washington State
The availability of substantially more genomic sequence data, revealing that WA2 belonged to a large, monophyletic ‘WA outbreak’ clade (17) , affords us the opportunity to revisit whether this outbreak actually began in January with WA1. This narrative has important implications not just for individuals and public health workers in Washington State associated with the WA1 case, but also for evaluating the effectiveness of state- and national-level mitigation strategies in the weeks following the first detection of SARS-CoV-2 in the US.
First, we noted that the ~3% probability reported by Bedford et al (4, 17) that WA1 and the WA outbreak clade would appear so close on the tree by chance is an underestimate: at least four transmission chains co-circulated in the state (their Figure 2), increasing the probability of sampling one that happened to have a close relationship to a rare lineage to >11%. Next, we note a peculiar feature of the relationship between WA1 and the WA outbreak clade. They differ by two substitutions, C17747T and A17858G, and despite hundreds of genomes sequenced in Washington State, no viruses with genomes identical to WA1 or transitional between it and the outbreak clade (i.e. having a C at position 17747 or an A at position 17858, like WA1) (17) had been sampled there. This absence was surprising given that scores of genomes identical to the putative descendant of WA1—and inferred most recent common ancestor (MRCA) of the WA outbreak clade—encoding both C17747T and A17858G, have been sampled from Feb 20th until April 27th (Supplementary text). Indeed, a range of phylogenetic patterns could have been observed (Fig. 1A–C), yet were not (Fig. 1D).
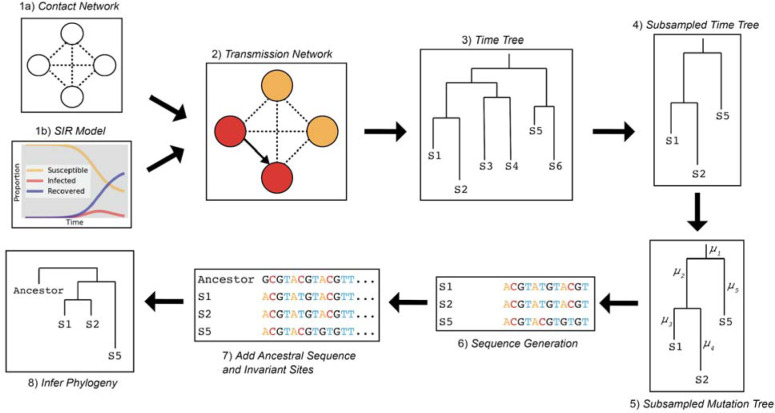
Fig. 2. Epidemic simulation workflow.
(1) FAVITES generates the contact network and (1a) runs an SIR model (1b) to simulate spread through the contact network and (2) produce a transmission network. (3) FAVITES outputs a viral time tree based on the infected individuals in the transmission network from which (4) individuals are subsampled to match the dates of the original epidemic (e.g., WA outbreak clade). (5) Evolutionary rates are applied to the time tree based on the number of variant sites from the original alignment, converting the branches from years to substitutions/site (μ). (6) Genetic sequences at variant sites are evolved over the subsampled tree using Pyvolve, starting with the ancestral sequence (e.g., WA1), based on GTR parameters inferred from the original alignment. (7) The ancestral sequence and invariant sites are added to the sequence data so (8) a maximum likelihood phylogeny can be inferred in IQ-TREE2.
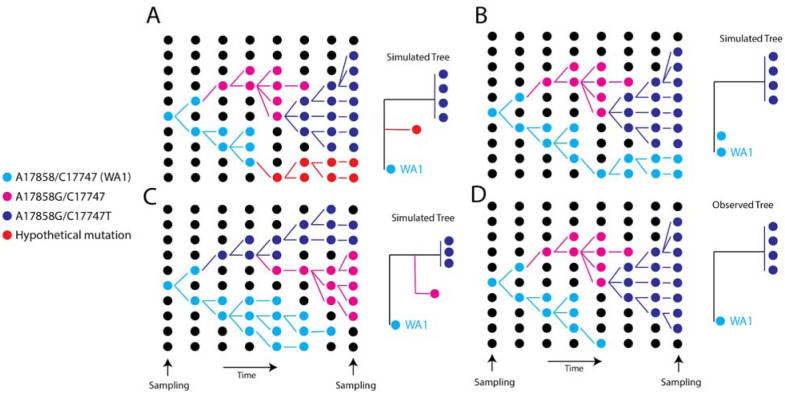
Fig. 1. Schematic showing a hypothetical path along which the key mutations in the WA outbreak could have taken in a susceptible population, alongside the inferred phylogeny.
(A) Scenario where a hypothetical mutation occurs from WA1-like genomes (B) A hypothetical phylogeny where A17747 and C17858 from the original WA1 virus are maintained in the population and sampled at the end. (C) Hypothetical scenario where a virus one mutation (A17747C) different from WA1 is maintained in the population. (D) The observed tree from the WA outbreak.
To investigate whether the observed pattern of evolution was consistent with the WA outbreak clade having descended from WA1, we simulated the entire WA outbreak clade, including the transmission history between thousands of cases, under realistic epidemiologic parameters (18), and then evolved genomes forward in time under the constraint that they originated from WA1 (Fig. 2). We simulated 1,000 epidemics seeded by WA1 on January 15th 2020 with a median doubling time of 4.7 days (95% range: 4.2–5.1) and an evolutionary rate of 0.8×10−3 substitutions/site/year. These simulated epidemics produced a median of 4,269 cases after 61 days (95% range: 1,993–11,053 cases). The median genetic distance from WA1 to the sub-sampled viruses was 3 mutations, which is consistent with the observed phylogeny.
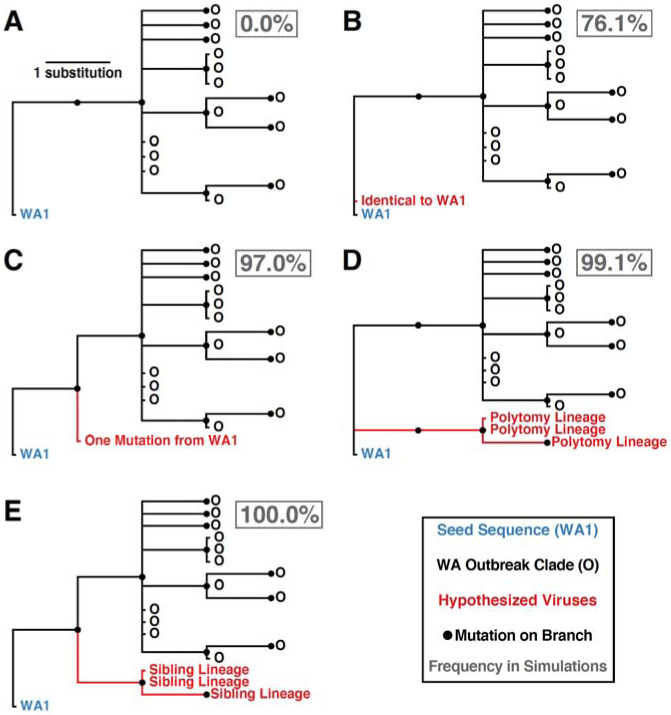
We examined the phylogenetic structure of maximum likelihood trees inferred from sub-sampled simulated viral sequences to determine how frequently they matched the observed relationship between WA1 and the WA outbreak clade. Specifically, a simulation tree matching the observed tree must produce a single branch emanating from WA1 that experiences at least two mutations (C17747T and A17858G in the observed tree) prior to establishment of a single outbreak clade (Fig. 3A). Alternative patterns include: (i) a virus identical to WA1 (Fig. 3B); (ii) a virus that differs from WA1 by a single mutation (Fig. 3C); (iii) a viral lineage forming a basal polytomy with WA1 and the outbreak clade (Fig. 3D); and (iv) a viral lineage that is sibling to the outbreak clade but only experienced a single mutation before divergence (Fig. 3E). The frequency of alternative phylogenetic patterns in the simulated epidemics represents the probability that the true topology (Fig. 3A) could not have occurred if the WA outbreak clade had been seeded by WA1.
Fig. 3. Potential phylogenetic relationships between WA1 and the Washington outbreak clade and their occurrence frequencies in 1000 epidemic simulations.
(A) Observed pattern where the WA1 genome is the direct ancestor of the outbreak clade, separated by at least two mutations. (B) Identical sequence to WA1. (C) Sequence that is one mutation divergent from WA1. (D) Lineage forming a basal polytomy with WA1 and the outbreak clade. (E) Sibling lineage to the outbreak clade experiencing only a single mutation from WA1 before divergence. Frequency each relationship was observed in 1000 simulations reported in gray box.
In 76.1% of simulations, we observed at least one virus genetically identical to WA1, with a median of 12 identical viruses in each simulation (95% range: 0–74 identical viruses). Not observing a virus identical to WA1 in the real Washington data does not significantly differ from expectation (p=0.239). However, viruses with one mutation from WA1 were observed in 97.0% of simulations, indicating a low probability of not detecting a single sequence from Washington within one mutation of WA1 (p=0.030). Lineages forming a basal polytomy with WA1 and the epidemic clade were observed in 99.1% of populations (p=0.009) and 100% of simulations had at least one sibling lineage diverging prior to the formation of the outbreak clade (p<0.001). Therefore, even if C17747T and A17858G were linked—a possibility since they are both non-synonymous mutations located in the nsp13 helicase gene—we would still expect to see descendants of their predecessors in Washington. In summary, when we seeded the Washington outbreak simulations with WA1 on January 15th, 2020, we failed to observe a single simulated epidemic that has the characteristics of the real phylogeny (Fig. 3). These findings are robust to simulations that used a slower epidemic doubling time of 5.6 days (95% range 5.2–5.9) or an accelerated mutation rate of 1.6×10−3 substitutions/site/year (18).
Accounting for geographical gaps in genomic sampling.
A major limitation in phylogeographic inference of SARS-CoV-2 to date has been the low availability of genomic sequence data from locations that experienced early outbreaks, including the original epicenter in Hubei Province, China. Although we do not have access to missing genome sequences, we can estimate how many such genomes are likely missing. We therefore developed methodologies that incorporate unsampled viruses into phylogeographic inferences within a Bayesian framework (18). We investigated how tree topologies were affected by the inclusion of unsampled viruses assigned to 13 of the most severely undersampled locations both in China and globally, based on COVID-19 incidence data (18). Realistic sampling time distributions also were inferred from incidence data. To better inform placement of unsampled viruses on the phylogeography, we developed a generalized linear model formulation that incorporates air passenger flows between regions within China and between Chinese regions and other countries.
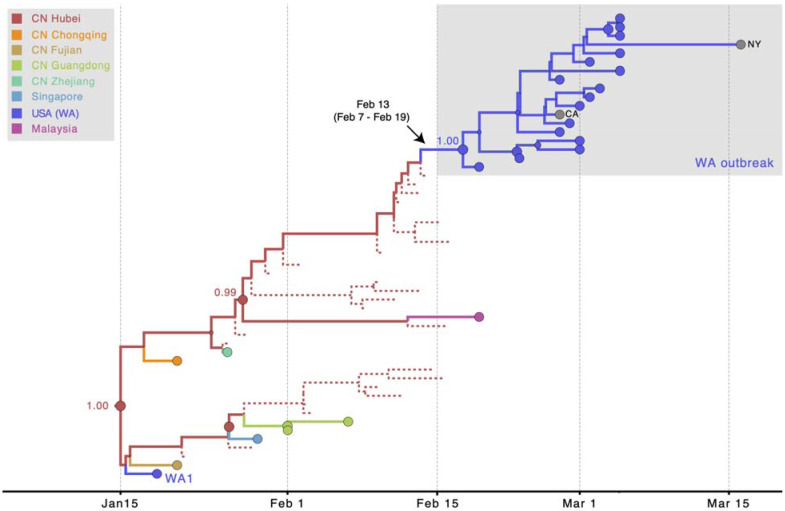
The resulting phylogeny (Fig. 4) provides a reconstruction of the evolutionary relationships of viruses from Washington State that realistically accounts for major gaps in sequence data from Hubei Province, China. For low-diversity data, a single MCC phylogeny has a resolution that is to a large extent not supported by the full posterior tree distribution, but key nodes yield good support in this case including their location estimates. The accommodation of unsampled viruses assigned to Hubei strongly supports independent introductions of WA1 and the WA outbreak clade from Hubei, with ancestral location states supported by high posterior probabilities (≥0.99). Furthermore, using Markov jump estimates that account for phylogenetic uncertainty (19), we inferred February 13th, 2020 (95% HPD February 7th–February 19th) as the time, along the branch leading up the MRCA of the WA outbreak clade, at which the founding virus of the WA outbreak clade arrived in Washington State from Hubei. Consistent with these estimates of the introduction date of this viral lineage into Washington State, the Seattle Flu Study tested 6,908 archived samples from January and February, of which only 10 from the end of February were positive (5).
Fig. 4. MCC tree of SARS-CoV-2 entry into Washington State.
A subtree of the maximum clade credibility (MCC) tree depicting the evolutionary relationships inferred between (i) the first identified SARS-CoV-2 case in the US (WA1); (ii) the clade associated with the Washington State outbreak (including WA2); and (iii) closely related viruses that were identified in multiple locations in Asia. Circles at the tips represent observed taxa and are shaded by location. Branches and internal node circles are shaded similarly by posterior modal location state. Dotted lines represent branches associated with unsampled taxa assigned to Hubei, China (CN). Circle sizes for internal nodes are proportional to posterior clade support. Posterior location state probabilities are shown for three well-supported key nodes.
This timing is approximately four weeks later than had been proposed (17), implying: (a) archived ‘self-swab’ samples may have retrospectively detected the virus within as little as a week of its arrival (5), (b) the Washington State outbreak may have been smaller than estimated based on the earlier inferences, and (c) the individual who introduced the founding virus likely arrived in the US after the initiation of the ‘Suspension of Entry’ of non-US residents from China on February 2nd, 2020 (20) but during the period when an estimated 40,000 US residents were repatriated from China, with screening described as cursory or lax (21). These passengers were directed to a short list of airports including Los Angeles, San Francisco, New York, Chicago, Newark, Detroit and Seattle (21). So, although our reconstructions incorporating unsampled lineages do not account for travel restrictions, the remaining influx likely provided ample opportunity for a second introduction to Washington State. It is also possible that the virus entered via nearby Vancouver, British Columbia, which is closely linked to both China and Washington State.
Early establishment of SARS-CoV-2 in Europe
Our simulation framework also suggested that the initial outbreak in Bavaria, Germany (represented by BavPat1) was unlikely to be responsible for seeding the Italian outbreak (see Fig. S1 for detailed phylogenetic scenarios). We simulated the origins of the Italian outbreak that was associated with viruses genetically related to BavPat1, again using realistic epidemiological parameters. Simulations with a median doubling time of 3.4 days (95% range: 2.9–4.4 days) resulted in a median epidemic size of 724.5 (95% range: 140–2,847) after 36 days. In the observed phylogeny, the Italian outbreak is the sole descendant lineage from BavPat1. Within the Italian outbreak, there are zero viruses identical to BavPat1 and four of the 27 related viruses included in this analysis are separated from BavPat1 by a single mutation. In simulation, the distributions of identical and one-mutation divergent viruses are not significantly different from expectation (p=0.146 and p=0.153, respectively). However, the lack of at least one descendent lineage that forms a polytomy with BavPat1 and the Italian outbreak significantly differs from expectation (p=0.005). Therefore, it is highly unlikely that BavPat1 or a virus identical to it seeded the Italian outbreak (Fig. S1).
An alternative scenario in which both the Germany and Italy outbreaks were independently introduced from China is further supported when missing sequences from undersampled locations are explicitly accommodated in the phylogeny. Using the approach described above, the evolutionary relationships of BavPat1 and viruses from the Italian outbreak were reconstructed while accommodating unsampled viruses assigned to Hubei and other locations (including Italy) determined to be undersampled, based on incidence data. The resulting phylogeny strongly supports independent viral introductions from Hubei into Germany and Italy, with the ancestral location states in Hubei supported by a posterior probability of 0.79 (Fig. 5). These findings again reveal that epidemiological linkages inferred from genetically similar SARS-CoV-2 viruses associated with outbreaks in different locations can be highly tenuous, given low levels of sampled viral genetic diversity and insufficient background data from key locations.
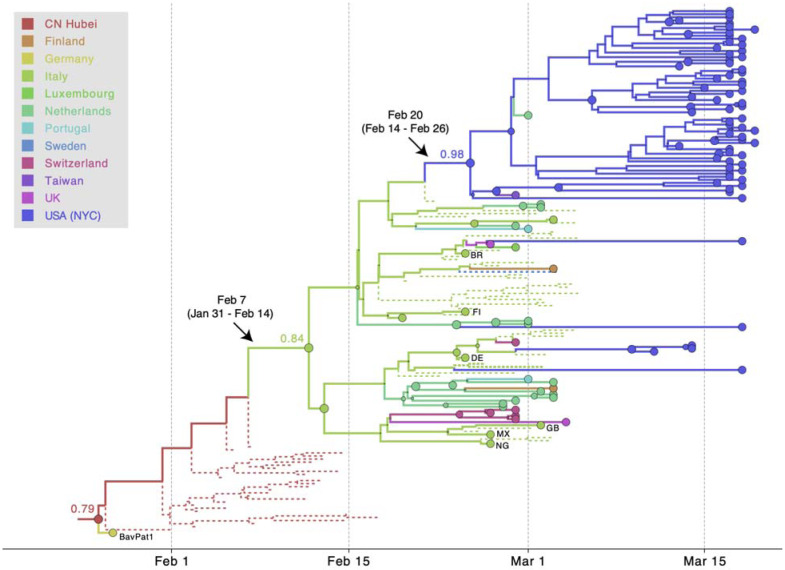
Fig. 5. MCC tree of SARS-CoV-2 entry into Europe.
A subtree inferred for viruses from (i) the first outbreak in Europe (Germany, BatPat1), (ii) outbreaks in Italy and New York, and (iii) other locations in Europe. Dotted lines represent branches associated with unsampled taxa assigned to Italy and Hubei, China (CN). Country codes are shown at tips for genomes sampled from travellers returning from Italy. Other features as described in Figure 4.
Interestingly, our approach is also able to infer that this major European clade, the same one that dominates in New York City (14, 15) and Arizona (22), had an origin in Italy, as might be expected from the epidemiological evidence. This inference makes use of the recent travel history, where available, of various sequences in that cluster from people who had recently travelled to Italy. There are only two samples available from Italy in that cluster, yet we can trace strong evidence of the origin of this important lineage to Italy via Hubei. The Markov jump estimates of the movement from Hubei to Italy were: Feb. 7th, 2020 (95% HPD Jan. 31st–Feb. 14th). This Italian/European cluster, in turn, was the source of multiple introductions to New York City (NYC) (14, 15). Using the same approach, we date the introduction leading to the largest NYC transmission cluster to Feb. 20th, 2020 (95% HPD Feb. 14th–Feb. 26th). Hence, even with its corrected age, the WA outbreak clade predates identified transmission clusters elsewhere in the US (22–24).
This global genomic perspective (Fig. 6) is relevant to recent claims (16) that this lineage is more transmissible due to the D614G Spike protein mutation. Instead, our results suggest the lineage possessing this mutation rode at least three early waves of uncontrolled transmission, first in the original Chinese epicenter of the outbreak, then in the earliest European epicenter in Northern Italy, and then in the uncontrolled outbreak in New York City. In other words, this viral lineage appears to have been amplified because of luck, not high fitness. We hypothesize that from these large centers of uncontrolled spread, where SARS-CoV-2 reached comparatively very high prevalence, D614G simply swamped D614 variants in other geographic regions (22, 25). While we await definitive evidence, what is clear is that sensational claims of functional importance of this mutation were premature.
Fig. 6. SARS-CoV-2 introductions to Europe and the US.
Pierce projection mapping early and apparently ‘dead-end’ introductions of SARS-CoV-2 to Europe and the US (dashed arrows). These were followed by a series of dispersals (solid arrows) all likely taking place in February 2020: from Hubei Province, China to Northern Italy, from Hubei to Washington State, then from Europe (as the Italian outbreak spread more widely) to New York City.
Discussion
January and February 2020 were pivotal months as government officials endeavored to understand and appropriately respond to the unfolding SARS-CoV-2 emergency, in the midst of considerable scientific uncertainty. When a new case was confirmed in a city that was not directly associated with travel, it was difficult to ascertain how the virus had gotten there and whether there had already been community transmission. There was also considerable uncertainty about whether epidemiological contact tracing and isolation would be effective for controlling new outbreaks. Weeks after the first diagnostic tests became available for SARS-CoV-2 on January 13th (26), the story of German public health workers successfully using testing, contact tracing and isolation to control a small outbreak in Germany had profound implications for the feasibility of early, intensive interventions to prevent the virus from becoming established in Germany and other countries (10). Assertions that these efforts had actually failed may have led to confusion about the utility of these approaches and contributed to a sense of the inevitability of the spread of the pandemic. Similarly, conclusions that the Seattle area was already six weeks into an epidemic by the end of February, rather than two or three, and the notion that stringent efforts to prevent spread had failed in the WA1 case, may have influenced decision-making about how to respond to the outbreak, including whether such measures were worth the effort.
Despite the early successes in containment, SARS-CoV-2 eventually took hold in both Europe and North America during February 2020: evidently first in Italy in early February, then in Washington State mid-February, and then in New York City later that month (Fig. 6). Our finding that the virus associated with the first known transmission network in the US did not enter the country until mid-February is sobering, since it demonstrates that the window of opportunity to block sustained transmission of the virus stretched all the way until that point. It is clear that early interventions can have outsized effects on the course of an outbreak, and the precise impact of the slow rollout of diagnostic tests in the US on the early stages of the pandemic is likely to be explored and debated for years to come, including the initially narrow criteria for who could be tested. Our findings critically inform such inquiries by delineating when community transmission was first established in the US and by providing clarity on the duration of the time window before SARS-CoV-2 establishment when contact tracing and isolation might have been most effective.
Our findings highlight the potential value of establishing intensive, community-level respiratory virus surveillance architectures, such as the Seattle Flu Study, during a pre-pandemic period. The value of detecting cases early, before they have bloomed into an outbreak, cannot be overstated in a pandemic situation (27). Given that every delay in case detection reduces the feasibility of containment, it is also worth assessing the impact of lengthy delays in FDA approval of testing the Seattle Flu Study’s stored samples for SARS-CoV-2.
By delaying COVID-19 outbreaks by even a few weeks in the US and Europe, the public health response to the WA1 case in Washington State, and a particularly impressive response in Germany to a substantial outbreak, bought crucial time for their own cities, as well as other countries and cities, to prepare for the virus when it finally did arrive. Erroneously suggesting that WA1 introduced the earliest US outbreak of SARS-CoV-2 obscured the societal and public health benefits produced by an attentive, collaborative, and thoughtful patient willing to work with public health workers to prevent the spread of SARS-CoV-2. One irony is the beneficial impact the decision of government officials in Washington State, to be among the first in the US to initiate restrictions on social distancing and size of gatherings, had even though the decision was founded at least in part on an assumption about the timing of community transmission not supported by the phylogenetic data (i.e. the belief that cryptic transmission had been ongoing since mid-January). This action may have closed the gap between the onset of sustained community transmission and mitigation measures in Washington State, compared to other locales like New York City, in ways that deserve careful reevaluation.
Because the SARS-CoV-2 evolutionary rate is slower than its transmission rate, many identical genomes are rapidly spreading. This genetic similarity places limitations on some inferences such as calculating the ratio of imported cases to local transmissions in a given area. Yet we have shown that, precisely because of this slow rate, when as little as one mutation separates viruses, this difference can provide enough information for hypothesis testing when appropriate methods are employed. Bearing this in mind will put us in a better position to understand SARS-CoV-2 in the coming years.
Supplementary Material
Acknowledgements:
We thank the patients and healthcare workers who made the collection of this global viral data set possible and all those who made viral genomic data available for analysis. We thank Niema Moshiri for his guidance on FAVITES.
Funding: MW was supported by the David and Lucile Packard Foundation as well as the University of Arizona College of Science and Office of Research Innovation and Impact. This work was supported by the Multinational Influenza Seasonal Mortality Study (MISMS), an on-going international collaborative effort to understand influenza epidemiology and evolution, led by the Fogarty International Center, NIH. The research leading to these results has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 725422-ReservoirDOCS ) and from the European Union’s Horizon 2020 project MOOD (grant agreement no. 874850). The Artic Network receives funding from the Wellcome Trust through project 206298/Z/17/Z. JOW acknowledges funding from the National Institutes of Health (K01AI110181, AI135992, and AI136056). PL acknowledges support by the Research Foundation -- Flanders (`Fonds voor Wetenschappelijk Onderzoek -- Vlaanderen’, G066215N, G0D5117N and G0B9317N). MAS acknowledges support from National Institutes of Health U19 AI135995. JBJ is thankful for support from the Canadian Institutes of Health Research Coronavirus Rapid Response Programme 440371 and Genome Canada for Bioinformatics and Computational Biology Programme 28PHY. JP acknowledges funding from the National Institutes of Health (T15LM011271). VH acknowledges funding from the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/M010996/1]. The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institutes of Health. We gratefully acknowledge support from NVIDIA Corporation with the donation of parallel computing resources used for this research.
Footnotes
Competing Interests: JOW has received funding from Gilead Sciences, LLC (completed) and the CDC (ongoing) via grants and contracts to his institution unrelated to this research. MAS receives funding from Janssen Research & Development, IQVIA and Private Health Management via contracts unrelated to this research.
Data and materials availability: BEAST .xml file example, FAVITES simulated phylogenies, and the GISAID accession numbers for all sequences used in the analysis are hosted at https://github.com/Worobeylab/SC2_outbreak
References and Notes:
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature. 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M. A., Weldon W. C., Biggs H. M., Uyeki T. M., Pillai S. K., Washington State 2019-nCoV Case Investigation Team, First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 382, 929–936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmon A., Inside the Race to Contain America’s First Coronavirus Case. The New York Times (2020), (available at https://www.nytimes.com/2020/02/05/us/corona-virus-washington-state.html).
- 4.T. B. (@trvrb), The team at the @seattleflustudy have sequenced the genome the #COVID19 community case reported yesterday from Snohomish County, WA, and have posted the sequence publicly to http://gisaid.org. There are some enormous implications here. Twitter (2020), (available at https://twitter.com/trvrb/status/1233970271318503426).
- 5.Chu H. Y., Englund J. A., Starita L. M., Famulare M., Brandstetter E., Nickerson D. A., Rieder M. J., Adler A., Lacombe K., Kim A. E., Graham C., Logue J., Wolf C. R., Heimonen J., McCulloch D. J., Han P. D., Sibley T. R., Lee J., Ilcisin M., Fay K., Burstein R., Martin B., Lockwood C. M., Thompson M., Lutz B., Jackson M., Hughes J. P., Boeckh M., Shendure J., Bedford T., Seattle Flu Study Investigators, Early Detection of Covid-19 through a Citywide Pandemic Surveillance Platform. N. Engl. J. Med. (2020), doi: 10.1056/NEJMc2008646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branswell H., Herper M., Feuerstein A., Silverman E., Florko N., Facher L., Washington State could see explosion in coronavirus cases, study says. STAT (2020), (available at https://www.statnews.com/2020/03/03/washington-state-risks-seeing-explosion-in-coronavirus-without-dramatic-action-new-analysis-says/).
- 7.Fink S., Baker M., Coronavirus May Have Spread in U.S. for Weeks, Gene Sequencing Suggests. The New York Times (2020), (available at https://www.nytimes.com/2020/03/01/health/coronavirus-washington-spread.html).
- 8.David Goodman J., How Delays and Unheeded Warnings Hindered New York’s Virus Fight. The New York Times (2020), (available at https://www.nytimes.com/2020/04/08/nyregion/new-york-coronavirus-response-delays.html).
- 9.Bolduc D. A., Webasto disputes link to Italy coronavirus outbreak. Automotive News (2020), (available at https://www.autonews.com/suppliers/webasto-disputes-link-italy-coronavirus-outbreak).
- 10.Böhmer M. M., Buchholz U., Corman V. M., Hoch M., Katz K., Marosevic D. V., Böhm S., Woudenberg T., Ackermann N., Konrad R., Eberle U., Treis B., Dangel A., Bengs K., Fingerle V., Berger A., Hörmansdorfer S., Ippisch S., Wicklein B., Grahl A., Pörtner K., Muller N., Zeitlmann N., Boender T. S., Cai W., Reich A., an der Heiden M., Rexroth U., Hamouda O., Schneider J., Veith T., Mühlemann B., Wölfel R., Antwerpen M., Walter M., Protzer U., Liebl B., Haas W., Sing A., Drosten C., Zapf A., Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect. Dis. (2020), doi: 10.1016/S1473-3099(20)30314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M., Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 382, 970–971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forster P., Forster L., Renfrew C., Forster M., Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 117, 9241–9243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rambaut A., Holmes E. C., Hill V., O’Toole Á., McCrone J. T., Ruis C., du Plessis L., Pybus O. G., A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv (2020), p. 2020.04.17.046086. [DOI] [PMC free article] [PubMed]
- 14.Maurano M. T., Ramaswami S., Westby G., Zappile P., Dimartino D., Shen G., Feng X., Ribeiro-dos-Santos A. M., Vulpescu N. A., Black M., Hogan M., Marier C., Meyn P., Zhang Y., Cadley J., Ordonez R., Luther R., Huang E., Guzman E., Serrano A., Belovarac B., Gindin T., Lytle A., Pinnell J., Vougiouklakis T., Boytard L., Chen J., Lin L. H., Rapkiewicz A., Raabe V., Samanovic-Golden M. I., Jour G., Osman I., Aguero-Rosenfeld M., Mulligan M. J., Cotzia P., Snuderl M., Heguy A., Sequencing identifies multiple, early introductions of SARS-CoV2 to New York City Region. medRxiv (2020), , doi: 10.1101/2020.04.15.20064931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Reiche A. S., Hernandez M. M., Sullivan M., Ciferri B., Alshammary H., Obla A., Fabre S., Kleiner G., Polanco J., Khan Z., Alburquerque B., van de Guchte A., Dutta J., Francoeur N., Melo B. S., Oussenko I., Deikus G., Soto J., Sridhar S. H., Wang Y.-C., Twyman K., Kasarskis A., Altman D. R., Smith M., Sebra R., Aberg J., Krammer F., Garcia-Sarstre A., Luksza M., Patel G., Paniz-Mondolfi A., Gitman M., Sordillo E. M., Simon V., van Bakel H., Introductions and early spread of SARS-CoV-2 in the New York City area. medRxiv, 2020.04.08.20056929 (2020). [DOI] [PMC free article] [PubMed]
- 16.Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E. E., Bhattacharya T., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., on behalf of the Sheffield COVID-19 Genomics Group, LaBranche C. C., Montefiori D. C., Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv (2020), p. 2020.04.29.069054.
- 17.Bedford T., Greninger A. L., Roychoudhury P., Starita L. M., Famulare M., Huang M.-L., Nalla A., Pepper G., Reinhardt A., Xie H., Shrestha L., Nguyen T. N., Adler A., Brandstetter E., Cho S., Giroux D., Han P. D., Fay K., Frazar C. D., Ilcisin M., Lacombe K., Lee J., Kiavand A., Richardson M., Sibley T. R., Truong M., Wolf C. R., Nickerson D. A., Rieder M. J., Englund J. A., the Seattle Flu Study Investigators, Hadfield J., Hodcroft E. B., Huddleston J., Moncla L. H., Müller N. F., Neher R. A., Deng X., Gu W., Federman S., Chiu C., Duchin J., Gautom R., Melly G., Hiatt B., Dykema P., Lindquist S., Queen K., Tao Y., Uehara A., Tong S., MacCannell D., Armstrong G. L., Baird G. S., Chu H. Y., Jerome K. R., Cryptic transmission of SARS-CoV-2 in Washington State. medRxiv (2020), , doi: 10.1101/2020.04.02.20051417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Materials and methods are available as supplementary materials at the Science website.
- 19.Minin V. N., Suchard M. A., Fast, accurate and simulation-free stochastic mapping. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 3985–3995 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proclamation on Suspension of Entry as Immigrants and Nonimmigrants of Persons who Pose a Risk of Transmitting 2019 Novel Coronavirus | The White House. The White House, (available at https://www.whitehouse.gov/presidential-actions/proclamation-suspension-entry-immigrants-nonimmigrants-persons-pose-risk-transmitting-2019-novel-coronavirus/).
- 21.Eder S., Fountain H., Keller M. H., Xiao M., Stevenson A., 430,000 People Have Traveled From China to U.S. Since Coronavirus Surfaced. The New York Times (2020), (available at https://www.nytimes.com/2020/04/04/us/coronavirus-china-travel-restrictions.html).
- 22.Ladner J. T., Larsen B. B., Bowers J. R., Hepp C. M., Bolyen E., Folkerts M., Sheridan K., Pfeiffer A., Yaglom H., Lemmer D., Sahl J. W., Kaelin E. A., Maqsood R., Bokulich N. A., Quirk G., Watt T. D., Komatsu K., Waddell V., Lim E. S., Caporaso J. G., Engelthaler D. M., Worobey M., Keim P., Defining the Pandemic at the State Level: Sequence-Based Epidemiology of the SARS-CoV-2 virus by the Arizona COVID-19 Genomics Union (ACGU). medRxiv (2020), , doi: 10.1101/2020.05.08.20095935 [DOI] [Google Scholar]
- 23.Deng X., Gu W., Federman S., Du Plessis L., Pybus O., Faria N., Wang C., Yu G., Pan C.-Y., Guevara H., Sotomayor-Gonzalez A., Zorn K., Gopez A., Servellita V., Hsu E., Miller S., Bedford T., Greninger A., Roychoudhury P., Famulare M., Chu H. Y., Shendure J., Starita L., Anderson C., Gangavarapu K., Zeller M., Spencer E., Andersen K., MacCannell D., Tong S., Armstrong G., Paden C., Li Y., Zhang Y., Morrow S., Willis M., Matyas B., Mase S., Kasirye O., Park M., Chan C., Yu A., Chai S., Villarino E., Bonin B., Wadford D., Chiu C. Y., A Genomic Survey of SARS-CoV-2 Reveals Multiple Introductions into Northern California without a Predominant Lineage. medRxiv (2020), , doi: 10.1101/2020.03.27.20044925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fauver J. R., Petrone M. E., Hodcroft E. B., Shioda K., Ehrlich H. Y., Watts A. G., Vogels C. B. F., Brito A. F., Alpert T., Muyombwe A., Razeq J., Downing R., Cheemarla N. R., Wyllie A. L., Kalinich C. C., Ott I. M., Quick J., Loman N. J., Neugebauer K. M., Greninger A. L., Jerome K. R., Roychoudhury P., Xie H., Shrestha L., Huang M.-L., Pitzer V. E., Iwasaki A., Omer S. B., Khan K., Bogoch I. I., Martinello R. A., Foxman E. F., Landry M. L., Neher R. A., Ko A. I., Grubaugh N. D., Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell (2020), doi: 10.1016/j.cell.2020.04.021 [DOI] [PMC free article] [PubMed]
- 25.Wagner C., Roychoudhury P., Hadfield J., Hodcroft E. B., Lee J., Moncla L. H., Müller N. F., Behrens C., Huang M.-L., Mathias P., Pepper G., Shrestha L., Xie H., Neher R. A., Baird G. S., Greninger A. L., Jerome K. R., Bedford T., Comparing viral load and clinical outcomes in Washington State across D614G mutation in spike protein of SARS-CoV-2, (available at https://github.com/blab/ncov-D614G).
- 26.Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR, (available at https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf).
- 27.Worobey M., Epidemiology: Molecular mapping of Zika spread. Nature. 546 (2017), pp. 355–357. [DOI] [PubMed] [Google Scholar]
- 28.Mossong J., Hens N., Jit M., Beutels P., Auranen K., Mikolajczyk R., Massari M., Salmaso S., Tomba G. S., Wallinga J., Heijne J., Sadkowska-Todys M., Rosinska M., Edmunds W. J., Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 5, e74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spielman S. J., Wilke C. O., Pyvolve: A Flexible Python Module for Simulating Sequences along Phylogenies. PLoS One. 10, e0139047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., Lanfear R., IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moshiri N., Ragonnet-Cronin M., Wertheim J. O., Mirarab S., FAVITES: simultaneous simulation of transmission networks, phylogenetic trees and sequences. Bioinformatics. 35, 1852–1861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayres D. L., Cummings M. P., Baele G., Darling A. E., Lewis P. O., Swofford D. L., Huelsenbeck J. P., Lemey P., Rambaut A., Suchard M. A., BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 68, 1052–1061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemey P., Rambaut A., Bedford T., Faria N., Bielejec F., Baele G., Russell C. A., Smith D. J., Pybus O. G., Brockmann D., Suchard M. A., Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2. PLoS Pathog. 10, e1003932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.