Abstract
Background. Severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) is the principal etiologic agent of SARS. We analyzed serum samples obtained from 623 patients with SARS in Beijing, to determine whether infection with SARS-CoV can elicit neutralizing antibodies (NAbs).
Methods. We developed a highly sensitive and safe neutralization assay using the SARS-CoV pseudotyped virus and used this assay to determine the titers of the NAbs in serum samples from patients with SARS.
Results. We found that 85.9% of serum samples contained NAbs against SARS-CoV and that most of the NAb activities could be attributed to immunoglobulin G. The NAbs became detectable first at 5–10 days after the onset of symptoms, and their levels peaked at 20–30 days and then were sustained for >150 days. The serum samples could neutralize the pseudotype particles bearing the spike glycoproteins from different SARS-CoV strains, suggesting that the NAbs to SARS-CoV were broadly reactive.
Conclusions. NAbs to SARS-CoV are broadly elicited in patients with SARS and, according to their kinetics, may correlate with viral load during the early stages of the disease. These results suggest that it is possible to develop effective vaccines against SARS and that NAbs provide a potential strategy for treating patients with SARS.
Severe acute respiratory syndrome (SARS) is a lifethreatening form of atypical pneumonia [1–3] that is caused by a newly identified coronavirus (CoV), SARS-associated CoV (SARS-CoV) [4–6]. Although the first epidemic of SARS is under control, its future reemergence is still possible. Therefore, effective strategies to control and treat this disease are urgently needed. Since SARS is an acute disease and the infection is naturally down-regulated during the early stages of the disease, host immunity against SARS-CoV appears to be effective. However, little is known about the immune control of SARS-CoV infection, especially the induced protective immune response in patients with SARS.
Neutralizing antibodies (NAbs) are key components in the protective immune responses to viral infections, because they can bind to viral particles and block them from entering the host cells [7–9]. Many CoVs are potent inducers of NAbs, which exhibit protective activities in vivo [10, 11]. The NAb against SARS-CoV has been analyzed recently, by use of a conventional neutralizing assay, in a small number of serum samples from patients with SARS [12]. Such a conventional assay requires culturing of wild-type virus, which is cumbersome and unsafe. In the present article, we describe an improved assay for analyzing NAbs to SARSCoV based on a newly established SARS-CoV pseudovirus system. This assay measures the expression of luciferase as a result of pseudovirus infection and is, therefore, highly sensitive and reproducible. In addition, the assay is safe and can be performed in a highthroughput fashion.
Using our neutralization assay, we analyzed serum samples obtained from 623 patients with SARS in the Beijing area, which was severely affected during the 2003 epidemic, with >25% of the total cases in the world. We found that a great majority (85.9%) of the serum samples from patients with SARS contained high titers of NAbs against SARS-CoV. We report here the characterization of the NAbs and the kinetics of generation of NAbs. Our results have important implications for the development of antibody therapy and vaccine for SARS.
Patients, Materials, and Methods
Serum samples. The patients with SARS described here were all probable cases, defined according to the Chinese Center for Disease Control and Prevention (CDC) case definition [13]. In brief, the patients were identified as having (1) a high fever, (2) acute respiratory distress syndrome, (3) chest radiographic findings of pneumonia, and (4) detection of SARS-CoV RNA by reverse-transcription polymerase chain reaction (RT-PCR).
Serum samples obtained from patients with SARS were provided by Ditan Hospital, Beijing (n=168), the Beijing Municipal CDC (n=93), and the Beijing Red Cross Blood Center (n=362). Sequential serum samples from 4 patients with SARS were provided by Peking Union Medical College Hospital. One hundred fifty-three normal serum samples were provided by the Campus Hospital of Peking University. To inactivate the complement in the serum samples, all samples were incubated in a water bath of 56°C for 30 min.
Preparation of SARS-CoV pseudotyped virus. The pHIVluc backbone plasmid (a gift from Dan R. Littman, New York University Medical Center, New York, NY) contains the HIV genome, with the envelope gene replaced by luciferase gene as a reporter. Plasmids (pc-TSh) were generated by cloning the synthetic and codon-optimized (“humanized”) spike (S) genes from strains BJ01, TOR2, Frankfurt, and TW1 into pcDNA 3.1+ (Invitrogen), as described elsewhere (Y. Nie, P. Wang, X. Shi, J. Chen, A. Zheng, W. Wang, X. Qu, G. Wang, M. Luo, L. Tan, X. Song, Z. Wang, X. Yin, J. Chen, M. Ding, and H. Deng, unpublished data). The pc-TSh and pHIV-luc plasmids were cotransfected into 293T cells by use of a calcium phosphate transfection protocol [14]. The supernatant was collected 48 h after transfection. The pseudotype virus was normalized by use of a p24 ELISA kit (bioMérieux) and was stored at +80°C.
Neutralization assay with pseudotype virus. A neutralization assay based on the pseudotype virus was performed by measuring the infection of Huh7 cells (a human hepatoma cell line) by use of the luciferase as a reporter gene [15]. In brief, Huh7 target cells were seeded in 96-well plates, at a density of 8×103 cells/well, and were incubated overnight at 37°C. The serum samples were serially diluted 2-fold, from 1:100 to 1: 3200 (or from 1:100 to 1:12,800 if the NAb ID50 was >3200), and were mixed with 0.5 ng (p24) of pseudotype virus. After incubation for 30 min at 37°C, the mixture was added to the target cells. Virus infectivity was determined 48 h later by measuring the amount of luciferase activity expressed in infected cells. Neutralization was calculated as an inhibition percentage of virus infection (luciferase activity) at each dilution, compared with the serum-negative controls, as follows: neutralization percentage=[1-(luciferase activity with serum/luciferase activity without serum)]×100 [16, 17]. The titers were expressed as the reciprocal of the final dilution of serum at the pseudovirus ID50, which was calculated by use of GraphPad Prism4 software (GraphPad Software).
Neutralization assay with live SARS-CoV. The SARS-CoV stock of BJ01 isolate was provided by the Chinese CDC. The serum samples were serially diluted 2-fold, from 1:20 to 1:640, and then were mixed with 100 TCID50 of the virus. After incubation for 1 h at 37°C, the mixture was inoculated onto 96- well plates of Vero cells. Cultures were held at 37°C and in 5% CO2, with daily microscopic examination for cytopathic effect (CPE). After a 3-day incubation, titers were expressed as the reciprocal of the highest dilution at which the CPE was completely inhibited [18].
Flow cytometric analysis. S protein-expressing HeLa (SHeLa) cells [19] and normal HeLa cells were collected and incubated for 30 min with the serum samples. After 3 washes with PBS plus 0.1% bovine serum albumin, the cells were incubated with the fluorescein isothiocyanate-conjugated goat anti-human IgG (1:100; Sigma) for 30 min. Finally, the cells were analyzed by use of a flow cytometer (MoFlo High-Performance Cell Sorter; Dako Cytomation).
ELISA. All serum samples were tested for SARS-CoV-specific antibodies by use of an indirect ELISA kit (BGI-GBI Biotech), which was coated with the lysate of SARS-CoV particles as antigens. All serum samples were diluted at 1:10, as recommended by the manufacturer. The optical density at 450 nm was measured on an ELISA plate reader (model 550; BioRad).
Serum IgG-related assay. The serum samples from patients with SARS were diluted in PBS and incubated with superfluous sepharose beads conjugated with staphylococcal protein A (SpA) to remove the IgG. Then the serum samples were tested for NAbs, as described above. The IgG from the serum samples from patients with SARS were purified with the SpA beads, in accordance with the manufacturer's protocol, and then were assayed for the neutralizing activity.
Results
Establishment of an assay for NAbs against SARS-CoV with pseudotype virus. To design a safe and sensitive assay for detection of NAbs to SARS-CoV, we generated an HIV pseudotype virus enveloped by the S protein from the BJ01 strain of SARS-CoV, with a luciferase gene as a reporter (HIV-luc/ SARS). This HIV-luc/SARS pseudovirus exhibited the highest infection level in Huh7 cells, which expressed a high level of the SARS-CoV receptor angiotensin-converting enzyme 2 [19]. This infection could be blocked by serum samples from patients with SARS and appeared to be SARS specific, because the same serum samples did not neutralize the vesicular stomatitis virus G glycoprotein pseudotyped virus (figure 1).
Figure 1.
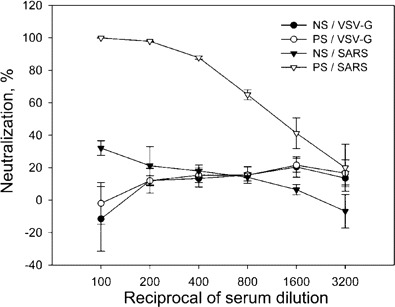
Detection of neutralizing antibodies in serum samples from a patient with severe acute respiratory syndrome (SARS) that can block the HIV-luc/SARS pseudovirus from entering and infecting Huh7 cells. Note the ability of the serum sample from the patient with SARS (PS) to neutralize and block the infectivity of HIV-luc/SARS (SARS); the absence of such blocking activity in normal serum (NS); and the lack of neutralizing activity of the SARS serum sample against the pseudotyped virus bearing the G protein of the vesicular stomatitis virus (VSV-G), which could infect the target cells at a level similar to that of HIV-luc/SARS. Data are the average of triplicate determinations. Bar, SD.
To evaluate the relevance of our pseudovirus neutralization assay, we randomly selected 10 serum samples from patients with SARS and 2 normal serum samples and analyzed them with the traditional neutralization assay, using live SARS-CoV, and then compared the results of 2 neutralization assays. As shown in table 1, the normal serum samples and 1 serum sample from a patient with SARS (D134) could not inhibit infection with either SARSCoV or the pseudovirus; the other 9 serum samples from patients with SARS showed neutralization activities in both assays.
To further validate the results from the neutralization tests, a binding assay was used to detect the S protein-specific antibodies in the above serum samples, with flow cytometric analysis using the S-HeLa cells (figure 2). Indications of positive antibodies were observed from the 9 serum samples from patients with SARS that also were positive in the neutralization tests. No S protein-specific antibodies were observed in the 2 normal serum samples and in serum sample D134. The consistency of the results in the binding assay and neutralization tests, with both the SARS-CoV and the pseudovirus, suggest that our pseudovirus-based neutralization assay is reliable for analyzing the S protein-specific NAbs in patients with SARS.
Figure 2.
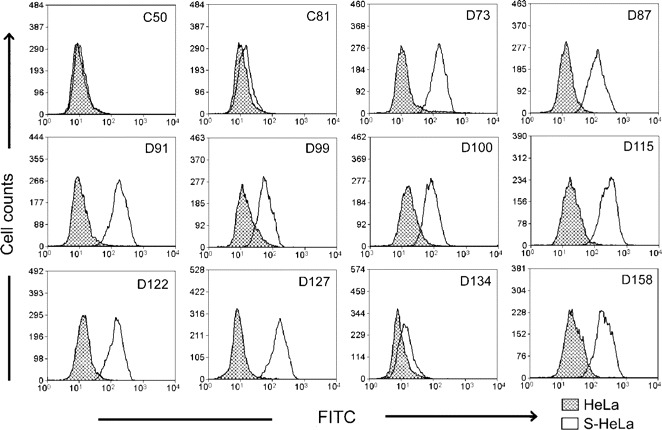
Detection of spike (S) protein-specific antibodies in serum samples, with flow cytometric analysis. C50 and C81 are 2 normal serum samples, and D73-D158 are 10 randomly selected serum samples from patients with severe acute respiratory syndrome. Data of flow cytometric analysis were analyzed with the Summit V3.1 program (Dako Cytomation). FITC, fluorescein isothiocyanate; HeLa, normal HeLa cells used as a control; S-HeLa, S protein-expressing HeLa cells.
NAbs in the serum samples from patients with SARS. To study the process of generation of NAbs, we first analyzed 153 normal human serum samples with our neutralization assay and, on the basis of the 99% confidence level (data not shown), set the negative/positive cutoff value at 329 ID50. We then analyzed the NAbs in sequential serum samples from 4 patients with wellcharacterized SARS cases (mean age, 34.5 years). Two of these patients progressed to serious atypical pneumonia, but none died (figure 3A). There were no NAbs detectable between days 5 and 8 after the onset of symptoms. From day 19 to day 30, all 4 patients generated NAbs with titers of >800 and maintained such high levels from day 33 to day 43. This result demonstrated that the NAbs could be induced in the patients with SARS and that the seroconversion of the S protein-specific NAbs occurred between days 8 and 19 after the onset of symptoms.
Figure 3.
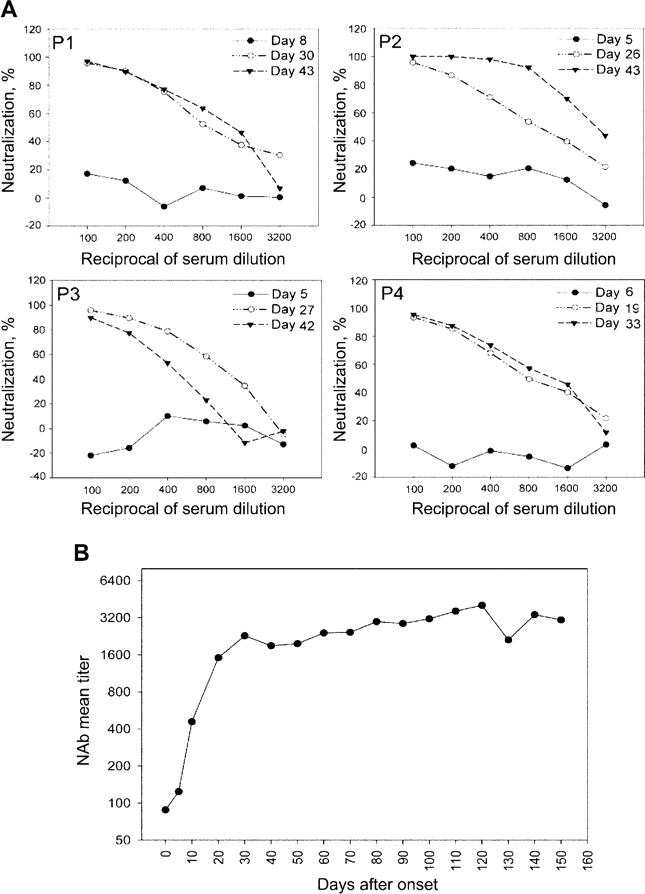
Kinetics of generation of neutralizing antibodies (NAbs) in serum samples obtained from patients with severe acute respiratory syndrome (SARS). A, Analysis of the NAbs in sequential serum samples obtained from 4 patients (P1, P2, P3, and P4). B, Analysis of NAbs in serum samples obtained from 623 patients with SARS. Each point on the curve contains 18 samples, and the entire curve contains data derived from the 623 serum samples. The titers of the NAbs, expressed in terms of ID50, are plotted against days after the onset of symptoms. The titer at day 0 is the mean ID50 (88) from 153 normal human serum samples.
To further define the kinetics of generation of NAbs, we analyzed a total of 623 serum samples from patients with SARS in Beijing. We found that 535 serum samples (85.9%) were positive (table 2), with an ID50 higher than the cutoff value of 329. The kinetics curve of the generation of NAbs (figure 3B), plotted against time after the onset of symptoms, showed that NAbs were first detectable from day 5 to day 10 after the onset of symptoms, reached a peak between days 20 and 30, and then remained at a high level (>2000) for at least 5 months.
Table 2.
Comparison of results of the neutralization assay and ELISA.
Cross-reactivity of the NAbs to different SARS-CoV strains. To evaluate the cross-reactivity of the NAbs in the serum samples from patients with SARS to different SARS-CoV strains, we randomly selected 168 of the 623 serum samples and analyzed their neutralizing activities against pseudotype viruses bearing the S protein from 4 different SARS-CoV strains (BJ01, Frankfurt, TOR2, and TW1). We found that all the serum samples were able to neutralize these 4 pseudotype viruses to similar extents. The representative results from 4 serumsamples are shown in figure 4.
Figure 4.
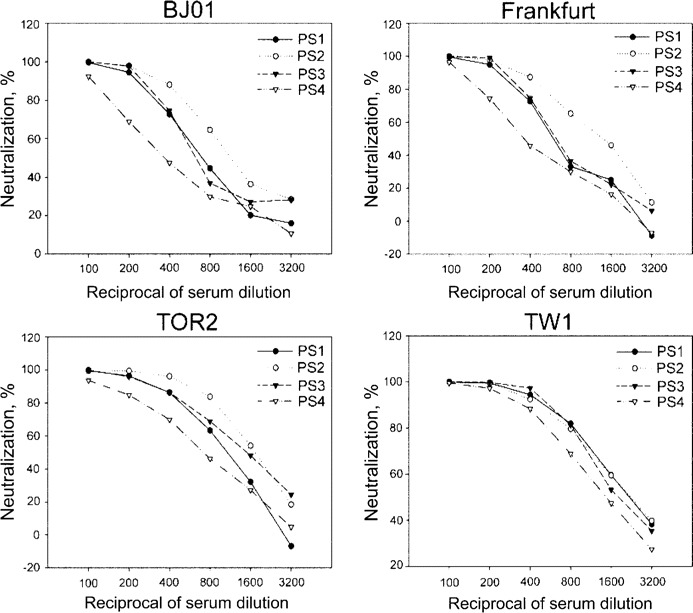
Neutralizing abilities of 4 serum samples from patients with severe acute respiratory syndrome (SARS) (PS1, PS2, PS3, and PS4) to the pseudotype viruses bearing the spike protein from 4 different SARS-associated coronavirus strains (BJ01, Frankfurt, TOR2, and TW1). Similar results were obtained in 3 independent experiments.
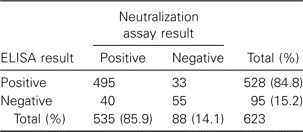
Comparison of the neutralization assay and ELISA. To compare our neutralization assay with other methods of serum analysis, we also analyzed the above 623 serum samples by use of a commercial ELISA kit, as described in Patients, Materials, and Methods.We found that 528 (84.8%) samples were positive by the ELISA. In addition, 495 (79.5%) samples were positive by both assays, and 55 (8.8%) were negative by both assays. Thirty-three samples were found to be neutralization negative but ELISA positive, and 40 samples were neutralization positive but ELISA negative (table 2). These results indicate that the neutralization assay and ELISA yielded consistent results in a great majority (88.3%) of the SARS cases.
Role of IgG in neutralization activity of serum samples from patients with SARS. To determine whether IgG plays a role in blocking the pseudovirus infection, we removed IgG from 3 positive serum samples by use of absorption with SpA beads. The IgG-depleted serum samples were found to lose most of their neutralizing activities (figure 5A). At the same time, the IgG fraction purified from 20 randomly selected serum samples from patients with SARS was able to significantly neutralize infection with the HIV-luc/SARS pseudovirus (figure 5B).
Figure 5.
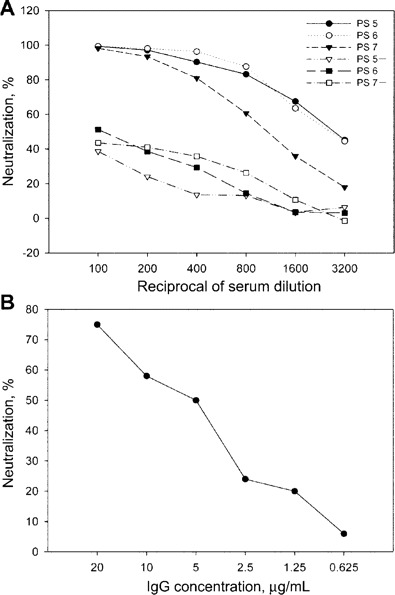
Neutralization caused by IgG in serum samples from patients with severe acute respiratory syndrome (SARS). A, Comparison of the neutralizing abilities of serum samples from patients with SARS, with or without IgG, using the neutralization assay. Serum samples from 3 patients (PS5, PS6, and PS7) were tested; “PS5+,” “PS6+,” and “PS7+ ”denote the same serum sample after IgG depletion. B, Neutralization assay of purified IgG from pooled positive serum samples. Similar results were obtained in 3 independent experiments
Discussion
Using a newly developed neutralization assay based on a SARS pseudotyped virus, we analyzed the NAbs to SARS-CoV present in a large population of patients with SARS, and we have described the kinetics of generation of NAbs in these patients (figure 3B). These kinetics were analogous to the profiles of the SARS-CoV-specific IgG reported by Peiris et al. [20] and Li et al. [21]. We also obtained evidence that much of the observed NAb activities are present in the purified IgG fraction (figure 5), indicating that IgG plays a major role in the neutralization of the SARS-CoV.
Our finding that the seroconversion of the S protein-specific NAbs happened between days 10 and 20 coincides with the decrease in the amount of SARS-CoV, as measured by RT-PCR of serum samples from patients with SARS during this period [20, 22, 23]. This result suggests that NAbs may play a role in the clearance of SARS-CoV during the early stages of the disease. A similar correlation has been observed in infections caused by other respiratory viruses. For example, Chargelegue et al. found that the NAbs to respiratory syncytial virus (RSV) were responsible for the reduction of the viral load in vivo [24]. Moreover, as reported in previous studies, the NAbs to RSV could be successfully used for treating high-risk infants with RSV infection [25], suggesting that the NAbs to SARS-CoV may have therapeutic potency at an early stage of the disease. Another important characteristic of the NAbs to SARS-CoV is that they are maintained at a high level for at least 5 months (figure 3B), which may provide a protective mechanism and an explanation for the remarkably low rate of SARS-CoV reinfection [26]. On the other hand, we found that there was no clear correlation between the NAb titer and the course of disease (data not shown), suggesting that the NAbs to SARS-CoV did not confer protection against the development of symptoms. This apparent contradiction can be explained, however, by a recent suggestion made by Peiris et al. [20] that the lung damage that occurs during the later stages of SARS may be caused by an overexuberant host immunological response, rather than uncontrolled replication of SARS-CoV [13].
Our NAb results are, in general, consistent (88.3%) with those obtained by use of a commercial ELISA kit (table 2). The inconsistency between these 2 assays may be due to the different antigens used and the sensitivity of these 2 assays. With regard to the 33 serum samples that were ELISA positive and neutralization negative, one possible reason for those results may be that the ELISA kit was coated with the SARS-CoV lysates and could detect the antibodies to other SARS-CoV structural proteins (including N, M, and E), as well as the S protein, whereas our neutralization assay could only detect the antibodies against the S protein. For the 40 neutralization-positive, ELISA-negative serum samples, to rule out false-positive results in our neutralization assay, we further analyzed 17 of the samples by use of the binding assay using the S-HeLa cells. Most of the samples (14 of 17) contained S protein-specific antibodies (data not shown), suggesting that, for the detection of S protein-specific NAbs, our neutralization assay was more sensitive than the ELISA kit.
In conclusion, our results demonstrate that SARS-CoV can elicit a strong humoral NAb response to the S protein, indicating that the S protein of this virus is highly immunogenic. In addition, our data show that NAb titers can be sustained for at least 150 days (figure 3B) and that the NAbs elicited by SARS-CoV S protein are not strain specific (figure 4). These findings provide strong support for the feasibility of developing a broadly reactive vaccine against SARS.
Table 1.
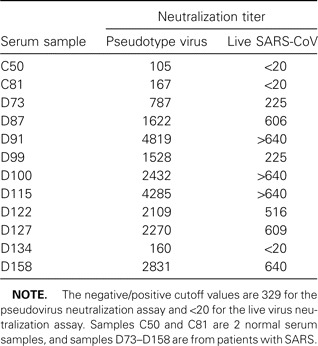
Titers of neutralizing antibodies in 12 serum samples analyzed with pseudovirus and live severe acute respiratory syndrome-associated coronavirus (SARS-CoV) neutralization assays.
Acknowledgements
We thank Tung-Tien Sun for critical reading of the manuscript and Yi Zha for helping to prepare a part of serum samples.
Footnotes
Financial support: National Nature Science Foundation of China (grant 30340027 and Outstanding Young Scientist Award 30125022 to H.D.); Ministry of Science and Technology (grant 2003CB514116 to H.D. and grant G1999011904 to M.D.); National Natural Science Foundation of China and German Research Association (grant GZ237 [202/10] to H.D.); 6th Framework Program of the European Commission (grant 511063 to H.D.).
References
- 1.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiel V, Ivanov KA, Putics A, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–15. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 3.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–70. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu YK, Hijikata M, Iwamoto A, Alter HJ, Purcell RH, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan M, Gawrieh S, Cotler SJ, Jensen DM. Neutralizing antibodies in hepatitis C virus infection: a review of immunological and clinical characteristics. Gastroenterology. 2003;125:597–604. doi: 10.1016/s0016-5085(03)00882-5. [DOI] [PubMed] [Google Scholar]
- 9.Crowe JE Jr, Suara RO, Brock S, Kallewaard N, House F, Weitkamp JH. Genetic and structural determinants of virus neutralizing antibodies. Immunol Res. 2001;23:135–45. doi: 10.1385/IR:23:2-3:135. [DOI] [PubMed] [Google Scholar]
- 10.Daniel C, Anderson R, Buchmeier MJ, et al. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J Virol. 1993;67:1185–94. doi: 10.1128/jvi.67.3.1185-1194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deregt D, Gifford GA, Ijaz MK, et al. Monoclonal antibodies to bovine coronavirus glycoproteins E2 and E3: demonstration of in vivo virusneutralizing activity. J Gen Virol. 1989;70:993–8. doi: 10.1099/0022-1317-70-4-993. [DOI] [PubMed] [Google Scholar]
- 12.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 13.Kamps BS, Hoffmann C. SARS reference. 3rd ed. Flying Publisher; 2003. Available at: http://www.sarsreference.com/sarsreference.pdf. Accessed 12 July 2004. [Google Scholar]
- 14.Deng H, Liu R, Ellmeier W, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H,Wang G, Li J, et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–45. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poignard P, Moulard M, Golez E, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77:353–65. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu HS, Chiu SC, Tseng TC, et al. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg Infect Dis. 2004;10:304–10. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Zheng A, Chen J, et al. Expression cloning of functional receptor used by SARS coronavirus. Biochem Biophys Res Commun. 2004;315:439–44. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia:a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–9. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization (WHO) Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) Geneva: WHO; 2003. Available at: http://www.who.int/entity/csr/sars/en/WHOconsensus.pdf. Accessed November 2003. [Google Scholar]
- 23.Gran PR, Garson JA, Tedder RS, Chan PK, Tam JS, Sung JJ. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med. 2003;349:2468–9. doi: 10.1056/NEJM200312183492522. [DOI] [PubMed] [Google Scholar]
- 24.Chargelegue D, Obeid OE, Hsu SC, et al. A peptide mimic of a protective epitope of respiratory syncytial virus selected from a combinatorial library induces virus-neutralizing antibodies and reduces viral load in vivo. J Virol. 1998;72:2040–6. doi: 10.1128/jvi.72.3.2040-2046.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.American Academy of Pediatrics Committee on Infectious Diseases and Committee of Fetus and Newborn Prevention of respiratory syncytial virus infections: indications for the use of palivizumab and update on the use of RSV-IGIV. Pediatrics. 1998;102:1211–6. doi: 10.1542/peds.102.5.1211. [DOI] [PubMed] [Google Scholar]
- 26.DeGroot AS. How the SARS vaccine effort can learn from HIV—speeding towards the future, learning from the past. Vaccine. 2003;21:4095–104. doi: 10.1016/S0264-410X(03)00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


