Abstract
The role of severe acute respiratory syndrome (SARS) coronavirus as an enteric pathogen was investigated in a cohort of 142 patients with SARS who were treated with a standard treatment protocol. Data from daily hematological, biochemical, radiological, and microbiological investigations were prospectively collected, and the correlation of these findings with diarrhea was retrospectively analyzed. Sixty-nine patients (48.6%) developed diarrhea at a mean (± standard deviation [SD]) of 7.6 ± 2.6 days after the onset of symptoms. The diarrhea was most severe at a mean (±SD) of 8.8 ± 2.4 days after onset, with a maximum frequency of 24 episodes per day (median, 5 episodes; range, 3–24 episodes). A higher mean virus load in nasopharyngeal specimens obtained on day 10 after the onset of symptoms was significantly associated with the occurrence of diarrhea (3.1 log10 vs. 1.8 log10 copies/mL; P = .01) and mortality (6.2 vs. 1.7 log10 copies/mL; P < .01). However, diarrhea was not associated with mortality. The lung and the gastrointestinal tract may react differently to SARS coronavirus infection. Additional investigation of the role of SARS coronavirus in the pathogenesis of diarrhea in patients with SARS should be conducted.
The severe acute respiratory syndrome (SARS) pandemic has affected >8000 patients, with 774 fatalities [1]. SARS is caused by a novel coronavirus, which was consistently isolated from patients with SARS who had subsequent seroconversion [2–4]. Similar histopathological findings of SARS and seroconversion have been reproduced in cynomolgus macaques (Macaca fascicularis) that were artificially inoculated with the same SARS coronavirus [5]. Subsequent detailed histopathological study of these infected macaques confirmed that the inflammatory changes were confined to the lungs [6]. Thus, SARS is largely regarded as a novel viral pneumonia. However, in a prospective clinical study of a cohort of patients with SARS, diarrhea was noted in 1% of patients at hospital admission and in 73% during hospitalization [7]. Viral genomes or the virus could be detected by RT-PCR or cell culture of stool samples obtained from these patients [8]. As illustrated by the example of enterovirus infection, shedding of virus in stool does not necessarily imply the presence of disease in the intestinal tract. Nevertheless, presence of numerous coronavirus particles was demonstrated in the terminal ileum and colon in a patient with SARS who had diarrhea [9]. It would be interesting to know whether the SARS coronavirus indeed causes diarrhea, because histopathological examination of intestinal biopsy specimens does not reveal any inflammatory or cytolytic damages.
We have previously shown that specimens obtained from the nasopharynx are useful for the diagnosis of rotavirus infection in children [10]. This is not unexpected, because an earlier study also demonstrated the presence of rotavirus in nasopharyngeal secretions from children with upper respiratory tract infection [11]. In this retrospective study, we attempt to correlate the virus load of SARS coronavirus shedding from the nasopharynx, the upper end of the aerodigestive tract, with the presence of diarrhea in a cohort of patients with SARS. Demographic characteristics, clinical features, and hematological and radiological findings of patients with SARS who had or did not have diarrhea were collected and analyzed.
Patients Materials and Methods
The clinical records for all 142 patients whose cases fulfilled the modified World Health Organization definition of SARS [4, 7] and who were treated at the United Christian Hospital and Caritas Medical Centre (Hong Kong) were analyzed. The daily clinical findings from history and physical examinations and hematological, biochemical, radiological, and microbiological investigations were prospectively collected and analyzed. In brief, the case definition includes fever (temperature, ⩾38°C), cough or shortness of breath, and new pulmonary infiltrates noted on chest radiographs or high-resolution CT, in the absence of an alternative diagnosis to explain the clinical presentation. Some of the clinical and virological results for the first 75 patients were previously reported [7]. All patients were treated with amoxicillin-clavulanate (1.2 g q8h iv) and azithromycin (500 mg q.d. po). In patients with a known allergy to penicillin, we administered 500 mg of oral levofloxacin every 24 h. As soon as the diagnosis of SARS was established, ribavirin (a 4-g oral loading dose, followed by 1.2 g q8h or 8 mg/kg q8h iv if the patient could not tolerate oral treatment) was given for 14 days, in addition to a tailing regimen of hydrocortisone (starting dose, 200 mg q8h iv) for 10 days, followed by oral prednisolone (1 mg/kg for 5 days, 0.5 mg/kg for 3 days, and 0.25 mg/kg for 3 days) for 11 days. Pulses of methylprednisolone (500–1000 mg q.d. iv for up to 3 g) were administered to patients with clinical deterioration. All hepatitis B surface antigen—positive patients were given prophylactic lamivudine (100 mg q.d. po) while taking corticosteroids.
Patients were prospectively monitored for development of diarrhea during hospitalization. Diarrhea was defined as ⩾3 bowel movements per day for ⩾2 consecutive days. The occurrence of diarrhea in relation to the onset of symptoms of SARS, the frequency of bowel movements, and the duration of diarrhea were recorded. Investigation for other causes of diarrhea was performed, including culture for Clostridium difficile, cell culture assay for detection C. difficile cytotoxin, and ELISA (IDEIA Rotavirus; DAKO) for detection of rotavirus.
For the diagnosis of coronavirus infection, nasopharyngeal specimens and serum samples were obtained at hospital admission. The convalescent-phase serum sample was taken 14–28 days after the onset of symptoms. For all patients, qualitative and quantitative RT-PCRs for SARS coronavirus were retrospectively performed using the nasopharyngeal specimens obtained at admission and 10 days after the onset of symptoms. Stool and urine specimens were obtained for RT-PCR during the hospital stay. All virological diagnostic protocols, including performance of qualitative and quantitative RT-PCRs, rapid viral antigen detection tests, viral cultures, immunofluorescent antibody tests for detection for IgG seroconversion against SARS-associated coronavirus, and other microbiological diagnostic evaluations, were done in the manner described in our previous publications [4, 7].
Statistical analysis. All data were calculated from the day of onset of clinical symptoms. We compared the demographic characteristics and laboratory values for patients with and without diarrhea by means of Fisher's exact test for categorical variables and Student's t test or the Mann-Whitney U test for continuous variables. A 2-tailed P value of <.05 was considered to be statistically significant. We used SPSS software, version 11.0 (SPSS), for all analyses.
Results
Of the 142 patients recruited in this study, 138 (97.2%) were ethnic Chinese, and the remaining patients were Filipinos. The patients were admitted to the hospital 2.4 ± 1.8 days after onset of symptoms. There were 56 male and 86 female patients (ratio, 0.65 : 1), and the mean age was 40.4 ± 13.9 years (range, 20–86 years). Eighty-six patients (60.6%) were household contacts of patients with SARS, and 28 (19.7%) were health care workers. For 28 patients (19.7%), the source of SARS infection could not be traced. Underlying chronic disease was found in 21 patients (14.8%), of whom 12 were chronic hepatitis B carriers without stigmata of chronic liver disease, 3 had diabetes mellitus, 3 had ischemic heart disease, 2 had malignancies, and 1 had chronic obstructive airway disease.
The clinical symptoms and signs of these 142 patients are shown in table 1. Common presenting symptoms included systemic upset, such as fever (96.5% of patients), chills (67.6%), and myalgia (57.7%). One-third of patients had tachycardia noted at admission. With regard to hematological and biochemical manifestations of disease, severe lymphopenia (lymphocyte count, <1 × 109 lymphocytes/L) was present in 82 patients (57.7%), and thrombocytopenia was present in 44 patients (31.0%) (table 2). Hyponatremia was noted in 27 patients (19%). Elevations in alanine aminotransferase, aspartate aminotransferase, creatinine kinase, and lactate dehydrogenase levels were noted in 27 (19.0%), 30 (21.1%), 36 (25.4%), and 39 (27.5%) of the patients, respectively. Initial chest radiograph findings were abnormal for 108 patients (76.1%), and multizone involvement was found in 29 of them (table 3). In 79 patients who presented with single-zone lesions, the right lower and left lower zones were more commonly involved than were the other zones. Thoracic CT was performed for 34 patients with initial apparently normal chest radiograph findings. The lesions were mostly confined to the retrocardiac region in the left lower lobes.
Table 1.
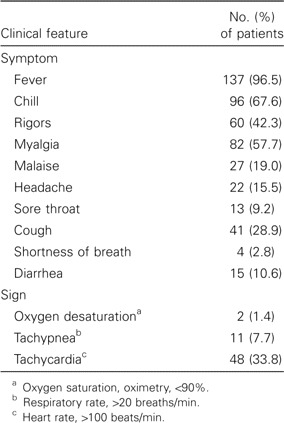
Clinical symptoms and signs for 142 patients with severe accurate respiratory syndrome at presentation.
Table 2.
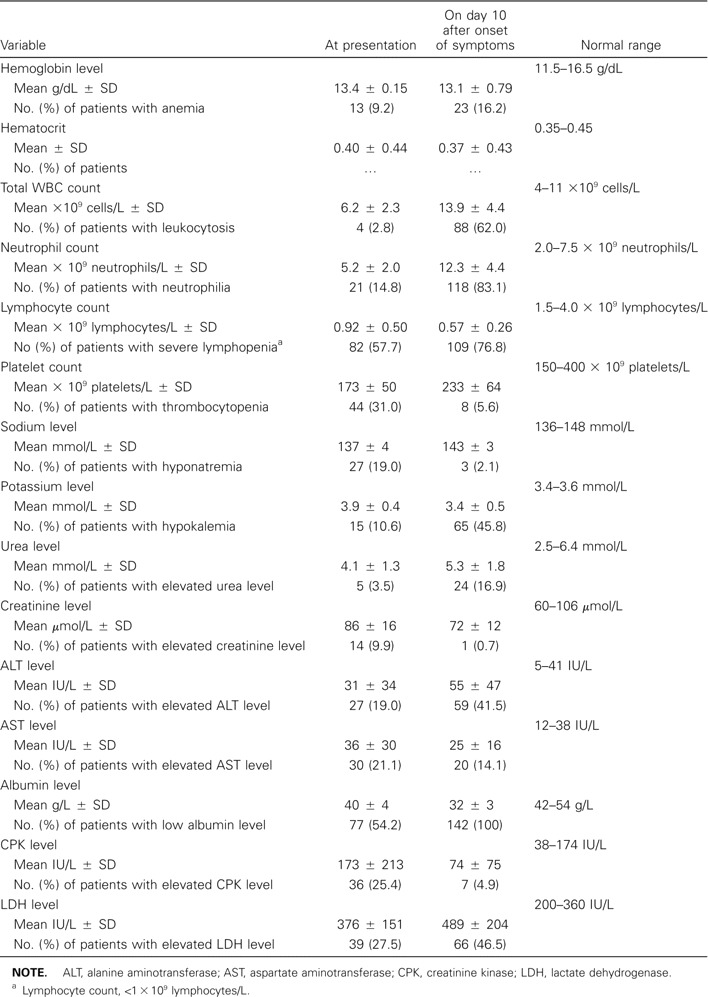
Hematological and biochemical findings for 142 patients with severe acute respiratory syndrome at presentation and on day 10 after the onset of symptoms.
Table 3.
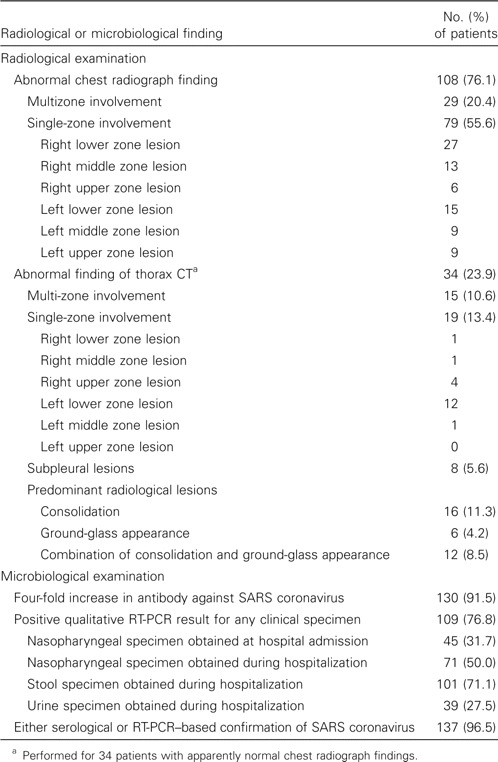
Extent of radiological involvement and microbiological findings for 142 patients with severe acute respiratory syndrome (SARS).
With regard to the microbiological evaluation for SARS coronavirus, 137 patients (96.5%) had either a 4-fold increase in antibody to SARS coronavirus or positive results of RT-PCR of nasopharyngeal, stool, or urine specimens (table 3). Four-fold increases in antibody titers to SARS coronavirus were found in 26 of 28 patients who did not have any history of epidemiological exposure, and RT-PCR results were positive for the remaining 2 patients.
During the course of infection, diarrhea occurred in 69 patients (48.6%). The distribution of episodes of diarrhea during the first 3 weeks of hospitalization is shown in figure 1. Diarrhea occurred 7.6 ± 2.6 days after the onset of symptoms and became most severe on day 8.8 ± 2.4, with a maximum frequency of 24 episodes per day (median, 5 episodes per day; range, 3–24 episodes per day). The diarrhea lasted for a duration of 3.8 ± 2.3 days in this cohort, resulting in prerenal azotemia in 6 patients and electrolyte disturbances in 39 patients. The diarrhea was rather painless, with no blood or mucus present in the stool for any patients. None of the patients had positive results of culture for C. difficile or cell culture assay for C. difficile cytotoxin. All stool samples were negative for rotavirus on ELISA tests.
Figure 1.
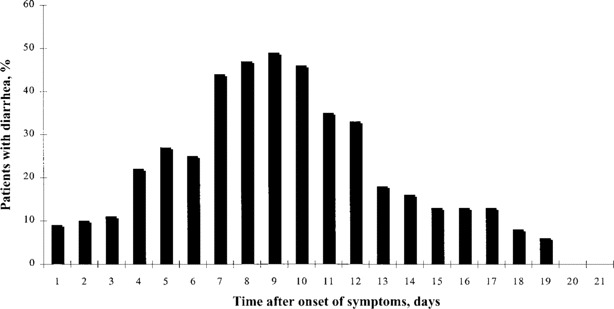
Percentage of patients with severe acute respiratory syndrome who had diarrhea during hospitalization
The clinical characteristics of patients with and those without diarrhea are summarized in table 4. There were no differences between the 2 groups with regard to age, sex, comorbidities, and chronic hepatitis B status. The manifestations on chest radiographs at hospital admission were also similar. There were no statistical differences in hematological and biochemical parameters between the 2 groups at presentation and on day 10 after the onset of symptoms. Patients with diarrhea had lower potassium levels (3.3 ± 0.5 mmol/L) and higher mean sodium levels (143 ± 3 mmol/L). There was no difference in the mean virus loads in nasopharyngeal specimens (as measured by quantitative RT-PCR) at baseline between the groups with and without diarrhea. A higher mean virus load in nasopharyngeal specimens obtained on day 10 after the onset of symptoms was significantly associated with the occurrence of diarrhea (3.1 vs. 1.8 log10 copies/mL; P = .01) and mortality (6.2 vs. 1.7 log10 copies/mL; P < .01). However, there were no differences between patients with and those without diarrhea with regard to the incidence of intensive care admission, receipt of mechanical ventilatory support, length of hospital stay, and overall mortality. Furthermore, the severity of diarrhea had no correlation with the virus load in nasopharyngeal specimens and overall mortality (figure 2).
Table 4.
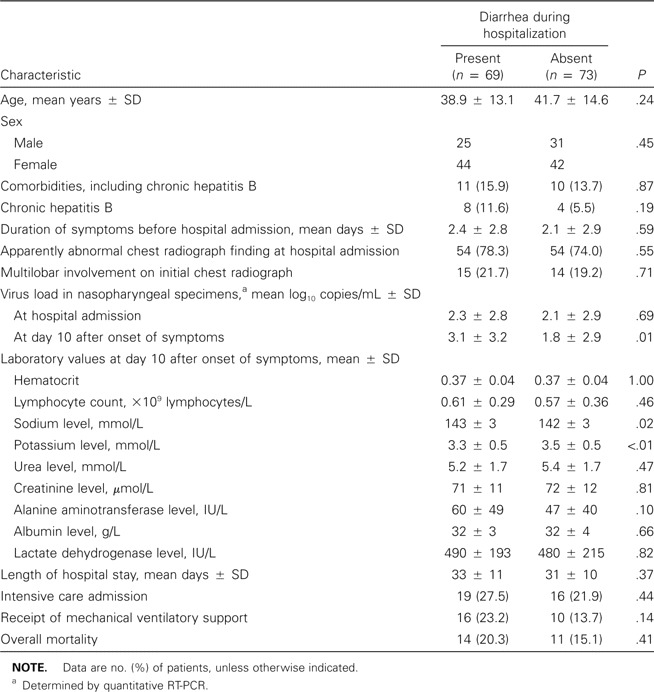
Demographic characteristics of 142 patients who had severe acute respiratory syndrome with or without diarrhea during hospitalization.
Figure 2.
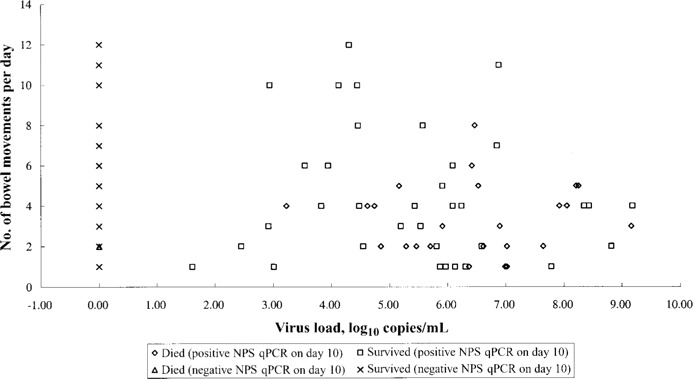
Frequency of bowel movements in relation to virus load in nasopharyngeal specimens on day 10 after the onset of symptoms. NPS, nasopharyngeal specimen; qPCR, quantitative RT-PCR.
Discussion
SARS is predominantly a viral pneumonia with a rapid tempo of deterioration. However, extrapulmonary manifestations are not uncommon. Skin rash [4], petechial cutaneous bleeding with prolonged activated partial thromboplastin time and increased d-dimer [12], impaired liver function, subclinical left ventricular diastolic dysfunction [13], and rhabdomyolysis [14] were occasionally reported. One of the more common presentations other than respiratory symptoms is diarrhea [4, 7, 9, 15–18]. Our previous report that >70% of patients in the Amoy Garden cohort developed diarrhea showed that fecal excretion may be an important mode of shedding and transmission [7]. Subsequent epidemiological investigation confirmed the importance of faulty sewage systems in propagating this massive outbreak of disease. Retrospective virological examination of stool samples by RT-PCR confirmed the presence of the virus in >90% of the samples obtained between day 14 and 21 after onset of symptoms, which is almost twice the positivity rate of the corresponding nasopharyngeal samples [8]. It is therefore interesting to examine the role of SARS coronavirus in the pathogenesis of diarrhea in patients with SARS.
Diarrhea is not an uncommon symptom during hospitalization for patients with infectious diseases other than acute gastroenteritis. In fact, diarrhea has been well reported in pneumonic illnesses other than SARS, such as legionnaires disease [19], Pneumocystis carinii pneumonia [20], and influenza (especially in children) [21]. Moreover, diarrhea is not uncommon in association with many systemic infections. For example, the rate of diarrhea was reported to be 34.5% among children with primary dengue fever [22]. The exact mechanisms leading to diarrhea in these systemic or pulmonary infections were not known, but they could be related to the cytokine profiles or metabolic changes associated with sepsis. In addition to primary viral illness, other potential causes of diarrhea include the use of drugs during hospitalization and C. difficile superinfection, because most patients with SARS have been empirically treated with antibiotics that cover typical and atypical pneumonia. Finally, diarrhea can be the direct result of viral cytolysis, because the virus can grow and produce a cytopathic effect in a large intestinal cell line, CACO-2 [23]. A study of the correlation between the clinical parameters, microbiological findings, and outcomes for patients with SARS with and without diarrhea may differentiate the relative importance of the suggested causative factors.
In this cohort of 142 patients with SARS, only 10.6% of the patients had diarrhea at hospital admission. During hospitalization, 69 (48.6%) of the patients had developed diarrhea at a mean time of 7.6 ± 2.6 days after the onset of symptoms. The most severe diarrhea occurred 1–2 days after the onset of diarrhea, with a maximum frequency of 24 episodes per day. In 6 patients, diarrhea was so severe that intravenous fluid hydration was warranted. The diarrhea was relatively painless and watery, as we have reported previously [7]. No correlation was found between diarrhea and hematological changes, radiological changes, use of antibacterials, or isolation of C. difficile and rotavirus from stool specimens. The only significant correlation was that there was a higher virus load in the nasopharyngeal specimens taken from patients with diarrhea on day 10 after the onset of symptoms. The finding that a higher virus load—but not the presence of diarrhea—predicted mortality in this group of patients is not unexpected. In 2 previous studies of animal models of SARS [6, 24], histological changes of acute respiratory distress syndrome (ARDS) and inflammation—characterized by diffuse alveolar damage (disruption of alveolar walls and infiltration with neutrophils and macrophages), alveolar hyaline membrane formation, as well as multinucleated syncytial cells—were found in a macaque monkey model artificially inoculated with SARS coronavirus [6]. None of the monkeys developed diarrhea over a period of 16 days, despite the fact that RT-PCR results were positive for SARS coronavirus in 1 of the stool samples. Similarly, no histopathological changes could be detected in the intestines of the monkeys, despite there being concomitant florid pathological changes in the lungs [6]. These findings suggest that viral replication triggers a different response and thus different functional impairment in the lungs and the gastrointestinal tract. A high virus load in the nasopharynx specimens either may predict severe pneumonia, ARDS, and mortality in some patients, or it may predict just diarrhea in other patients. This postulation is in keeping with the lack of significant difference in hematological markers of inflammation or increased cell turnover (lactate dehydrogenase level) between the groups with or without diarrhea. Although suppression of inflammatory changes in the intestine resulting from steroid therapy is possible, the routine use of steroids and ribavirin as part of the treatment protocol did not allow us to control the effect of use of steroids or antivirals on diarrhea. Moreover, the use of steroids has not affected the severe inflammatory reactions in the lungs of persons who have died of SARS [25].
In our previous study, we showed that rotavirus can be detected by viral isolation in cell culture and by indirect immunofluorescent antigen detection [10]. This is not unexpected, because many viruses that affect the gastrointestinal tract also affect the respiratory tract, and vice versa. For instance, in several studies, rotavirus has been well reported to be associated with bronchiolitis and fatal pneumonitis in immunocompromised as well as immunocompetent hosts [11, 26, 27]. Others reported that respiratory symptoms were found in 93% of patients with gastroenteritis caused by enteric adenovirus [28]. This virus has been found in samples from the upper respiratory tract and in stool samples obtained from these patients [29]. Similar to rotavirus and adenovirus, the SARS coronavirus can multiply and shed in the mucosa of the upper aerodigestive tract. In this regard, determination of the virus load in specimens from the nasopharynx may be useful for predicting the severity of illness in both the respiratory and the gastrointestinal tracts. From the laboratory perspective, RT-PCR is more likely to be affected by the abundant inhibitors present in stool than by nasopharyngeal secretions. Moreover, the volume of diarrhea varies markedly from patient to patient. This may also affect the interpretation of data on the virus load in stool. Therefore, we attempted to use nasopharyngeal specimens instead of stool specimens in this study.
Coronaviruses can cause respiratory or diarrheal diseases in both human and animals [30–44]. Human enteric coronavirus has been seen in diarrheal stool samples, but culture methods and characterization are still problematic [30, 31]. Human coronavirus OC43 and 229E are known to cause one-third of cases of the common cold [45, 46]. In the case of porcine coronavirus, a collection of transmissible gastroenteritis coronavirus (TGEV) recombinants were generated to study the molecular basis of TGEV tropism [47]. Recombinants of group 1 had enteric and respiratory tropism, whereas group 2 recombinants infected the respiratory but not the enteric tract. A substitution in amino acid 219 of the S protein in group 1 recombinant was responsible for the loss of enteric tropism [47]. On the other hand, targeted recombination of the S gene of TGEV resulted in a change from respiratory to enteric tropism and enhanced virulence [48]. However, no major deletion or mutation of functional significance had been identified by genomic sequencing of SARS coronavirus isolated from the diarrheal patients [49].
We have clearly demonstrated the presence of a numerous virus in tissue biopsy specimens from the terminal ileum and colon of a SARS patient with diarrhea [9]. The virus is capable of active replication inside the endoplasmic reticulum and adheres to the surface of enterocytes in both small and large intestines (as shown by electron microscopy), without causing significant villous atrophy or inflammatory cell infiltration in the mucosal or submucosal regions [9]. The absence of inflammation, cell necrosis, or microvilli atrophy does not exclude SARS coronavirus as the cause of diarrhea in these patients. For astrovirus-associated diarrhea in turkeys, the animal model has demonstrated mild histological changes, with a surprising lack of inflammation due to increased activation of the potent immunosuppressive cytokine transforming growth factor β at the peak of diarrhea during astrovirus infection [50]. Additional studies should be performed to ascertain the mechanism used by SARS coronavirus in the pathogenesis of diarrhea in humans.
References
- 1.World Health Organization . Summary table of SARS cases by country, 1 November 2002–7 August 2003. 23 September. 2003. Available at: http://www.who.int/csr/sars/country/en/country2003_08_15.pdf. [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier RA, Kuiken T, Schutten M, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–70. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KH, Poon LLM, Cheng VCC, et al. Emerg Infect Dis. Detection of SARS coronavirus (SCoV) by RT-PCR, culture, and serology in patients with acute respiratory syndrome (SARS) (in press) [Google Scholar]
- 9.Leung WK, To KF, Chan PK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–7. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng BJ, Chang RX, Ma GZ, et al. Rotavirus infection of the oropharynx and respiratory tract in young children. J Med Virol. 1991;34:29–37. doi: 10.1002/jmv.1890340106. [DOI] [PubMed] [Google Scholar]
- 11.Fragoso M, Kumar A, Murray DL. Rotavirus in nasopharyngeal secretions of children with upper respiratory tract infection. Diagn Microbiol Infect Dis. 1986;4:87–8. doi: 10.1016/0732-8893(86)90062-3. [DOI] [PubMed] [Google Scholar]
- 12.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–94. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 13.Li SS, Cheng CW, Fu CL, et al. Left ventricular performance in patients with severe acute respiratory syndrome: a 30-day echocardiogram follow-up study. Circulation. 2003;108:1798–803. doi: 10.1161/01.CIR.0000094737.21775.32. [DOI] [PubMed] [Google Scholar]
- 14.Wang JL, Wang JT, Yu CJ, et al. Rhabdomyolysis associated with probable SAR. Am J Med. 2003;115:421–2. doi: 10.1016/S0002-9343(03)00448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan JW, Ng CK, Chan YH, et al. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 2003;58:686–9. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Z, Zhang F, Xu M, et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52:715–20. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 17.Hsu LY, Lee CC, Green JA, et al. Severe acute respiratory syndrome (SARS) in Singapore: clinical features of index patient and initial contact. Emerg Infect Dis. 2003;9:713–7. doi: 10.3201/eid0906.030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto are. JAMA. 2003;289:2801–9. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 19.Sopena N, Sabria-Leal M, Pedro-Botet ML, et al. Comparative study of the clinical presentation of Legionella pneumonia and other community-acquired pneumonia. Chest. 1998;113:1195–200. doi: 10.1378/chest.113.5.1195. [DOI] [PubMed] [Google Scholar]
- 20.Newton JA, Jr, Weiss PJ, Litynski JJ. Diarrhea in Pneumocystis carinii pneumonia. Am J Gastroenterol. 1993;88:1138–9. [PubMed] [Google Scholar]
- 21.Wang YH, Huang YC, Chang LY, et al. Clinical characteristics of children with influenza A virus infection requiring hospitalization. J Microbiol Immunol Infect. 2003;36:111–6. [PubMed] [Google Scholar]
- 22.Pancharoen C, Mekmullica J, Thisyakorn U. Primary dengue infection: what are the clinical distinctions from secondary infection. Southeast Asian J Trop Med Public Health. 2001;32:476–80. [PubMed] [Google Scholar]
- 23.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferon. Lancet. 2003;362:293–4. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martina BE, Haagmans BL, Kuiken T, et al. Virology: SARS virus infection of cats and ferret. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–8. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouard J, Freymuth F, Vabret A, Jokic M, Guillois B, Duhamel JF. Viral co-infections in immunocompetent infants with bronchiolitis: prospective epidemiologic study. Arch Pediatr. 2000;7(3):531–5. doi: 10.1016/s0929-693x(00)80180-3. [DOI] [PubMed] [Google Scholar]
- 27.Nuovo GJ, Owor G, Andrew T, Magro C. Histologic distribution of fatal rotaviral pneumonitis: an immunohistochemical and RT in situ PCR analysis. Diagn Mol Pathol. 2002;11:140–5. doi: 10.1097/00019606-200209000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Yolken RH, Lawrence F, Leister F, Takiff HE, Strauss SE. Gastroenteritis associated with enteric type adenovirus in hospitalized infant. J Pediatr. 1982;101:21–6. doi: 10.1016/s0022-3476(82)80173-x. [DOI] [PubMed] [Google Scholar]
- 29.Brandt CD, Kim HW, Rodriguez WJ, Arrobio JO, Jeffries BC, Parrott RH. Simultaneous infections with different enteric and respiratory tract viruse. J Clin Microbiol. 1986;23:177–9. doi: 10.1128/jcm.23.1.177-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resta S, Luby JP, Rosenfeld CR, Siegel JD. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–81. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- 31.Gerna G, Passarani N, Battaglia M, Rondanelli EG. Human enteric coronaviruses: antigenic relatedness to human coronavirus OC43 and possible etiologic role in viral gastroenteriti. J Infect Dis. 1985;151:796–803. doi: 10.1093/infdis/151.5.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavallaro JJ, Monto AS. Community-wide outbreak of infection with a 229E-like coronavirus in Tecumseh, Michigan. J Infect Dis. 1970;122:272–9. doi: 10.1093/infdis/122.4.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185:1338–41. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 2003;36:985–9. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang XM, Herbst W, Kousoulas KG, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic chile. J Med Virol. 1994;44:152–61. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark MA. Bovine coronavirus. Br Vet J. 1993;149:51–70. doi: 10.1016/S0007-1935(05)80210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laude H, Van Reeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interaction. Vet Res. 1993;24:125–50. [PubMed] [Google Scholar]
- 38.Gelinas AM, Boutin M, Sasseville AM, Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 2001;76:43–57. doi: 10.1016/S0168-1702(01)00243-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsunemitsu H, el-Kanawati ZR, Smith DR, Reed HH, Saif LJ. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J Clin Microbiol. 1995;33:3264–9. doi: 10.1128/jcm.33.12.3264-3269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochizuki M, Osawa N, Ishida T. Feline coronavirus participation in diarrhea of cat. J Vet Med Sci. 1999;61:1071–3. doi: 10.1292/jvms.61.1071. [DOI] [PubMed] [Google Scholar]
- 41.Cho KO, Halbur PG, Bruna JD, et al. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J Am Vet Med Assoc. 2000;217:1191–4. doi: 10.2460/javma.2000.217.1191. [DOI] [PubMed] [Google Scholar]
- 42.Williams BH, Kiupel M, West KH, Raymond JT, Grant CK, Glickman LT. Coronavirus-associated epizootic catarrhal enteritis in ferret. J Am Vet Med Assoc. 2000;217:526–30. doi: 10.2460/javma.2000.217.526. [DOI] [PubMed] [Google Scholar]
- 43.Wunschmann A, Frank R, Pomeroy K, Kapil S. Enteric coronavirus infection in a juvenile dromedary (Camelus dromedarius. J Vet Diagn Invest. 2002;14:441–4. doi: 10.1177/104063870201400518. [DOI] [PubMed] [Google Scholar]
- 44.Ignjatovic J, Sapats S. Avian infectious bronchitis virus. Rev Sci Tech. 2000;19:493–508. doi: 10.20506/rst.19.2.1228. [DOI] [PubMed] [Google Scholar]
- 45.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infection. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heikkinen T, Jarvinen A. The common cold. Lancet. 2003;361:51–9. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballesteros ML, Sanchez CM, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropis. Virology. 1997;227:378–88. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez CM, Izeta A, Sanchez-Morgado JM, et al. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 1999;73:7607–18. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan Y, Peiris JS, Zheng B, et al. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:88104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koci MD, Moser LA, Kelley LA, Larsen D, Brown CC, Schultz-Cherry S. Astrovirus induces diarrhea in the absence of inflammation and cell death. J Virol. 2003;77:11798–808. doi: 10.1128/JVI.77.21.11798-11808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


