Zoonotic diseases present a substantial global health burden. In this Opinion article, Plowrightet al. present an integrative conceptual and quantitative model that reveals that all zoonotic pathogens must overcome a hierarchical series of barriers to cause spillover infections in humans.
Supplementary information
The online version of this article (doi:10.1038/nrmicro.2017.45) contains supplementary material, which is available to authorized users.
Subject terms: Diseases, Policy and public health in microbiology, Pathogens
Abstract
Zoonotic spillover, which is the transmission of a pathogen from a vertebrate animal to a human, presents a global public health burden but is a poorly understood phenomenon. Zoonotic spillover requires several factors to align, including the ecological, epidemiological and behavioural determinants of pathogen exposure, and the within-human factors that affect susceptibility to infection. In this Opinion article, we propose a synthetic framework for animal-to-human transmission that integrates the relevant mechanisms. This framework reveals that all zoonotic pathogens must overcome a hierarchical series of barriers to cause spillover infections in humans. Understanding how these barriers are functionally and quantitatively linked, and how they interact in space and time, will substantially improve our ability to predict or prevent spillover events. This work provides a foundation for transdisciplinary investigation of spillover and synthetic theory on zoonotic transmission.
Supplementary information
The online version of this article (doi:10.1038/nrmicro.2017.45) contains supplementary material, which is available to authorized users.
Main
The phenomenon of cross-species spillover is the defining characteristic of pathogens that transmit from vertebrate animals to humans (zoonoses). The public health burden that is presented by zoonoses includes outbreaks of pathogens such as Ebola virus, influenza A virus (H1N1)pdm09 and Middle East respiratory syndrome coronavirus (MERS-CoV), as well as the ongoing transmission of endemic pathogens, such as Salmonella spp., Leptospira spp., Trypanosoma spp., Mycobacterium spp. and West Nile virus1,2,3,4,5,6.
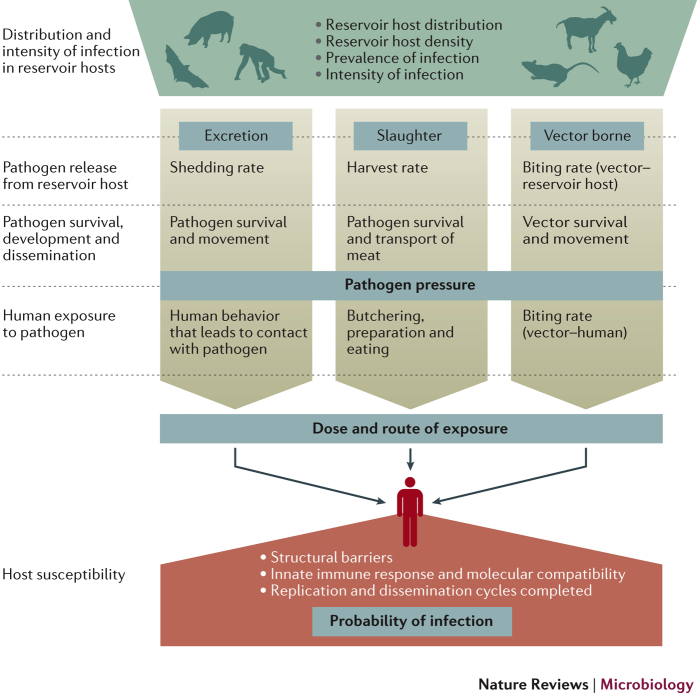
Spillover transmission is promoted by successive processes that enable an animal pathogen to establish infection in a human. The probability of zoonotic spillover is determined by interactions among several factors, including disease dynamics in the reservoir host, pathogen exposure and the within-human factors that affect susceptibility to infections. These factors can be partitioned into three functional phases that describe all major routes of transmission (Fig. 1). In the first phase, the amount of pathogen available to the human host at a given point in space and time, known as the pathogen pressure, is determined by interactions among reservoir host distribution, pathogen prevalence and pathogen release from the reservoir host, followed by pathogen survival, development and dissemination outside of the reservoir hosts. Second, human and vector behaviour determine pathogen exposure; specifically, the likelihood, route and dose of exposure. Third, genetic, physiological and immunological attributes of the recipient human host, together with the dose and route of exposure, affect the probability and severity of infection.
Figure 1. Pathways to spillover.
The risk of spillover is determined by a series of processes that link the ecological dynamics of infection in reservoir hosts, the microbiological and vector determinants of survival and dissemination outside of reservoir hosts, the epidemiological and behavioural determinants of exposure, and the within-host biological factors that shape the susceptibility of recipient hosts. The distribution and intensity of infection in reservoir hosts, followed by pathogen release, movement, survival and possible development to infectious stage, determine the pathogen pressure, which is defined as the amount of pathogen available to the recipient host at a given point in space and time. Pathogen pressure then interacts with the behaviour of the recipient host (and vector for vector-borne pathogens) to determine the likelihood, dose and route of exposure. A series of within-host barriers then determine host susceptibility, and, therefore, the probability and severity of infection for a given pathogen dose.
Each phase presents multiple barriers to the flow of a pathogen from a reservoir host to a recipient host. Spillover requires the pathogen to pass every barrier and thus can only occur when gaps align in each successive barrier within an appropriate window in space and time (Fig. 2). Consequently, zoonotic spillover is a relatively rare event, and although humans are continually exposed to many potentially infectious pathogens that are derived from other species, most of these microorganisms cannot infect or cause disease in humans7,8,9,10.
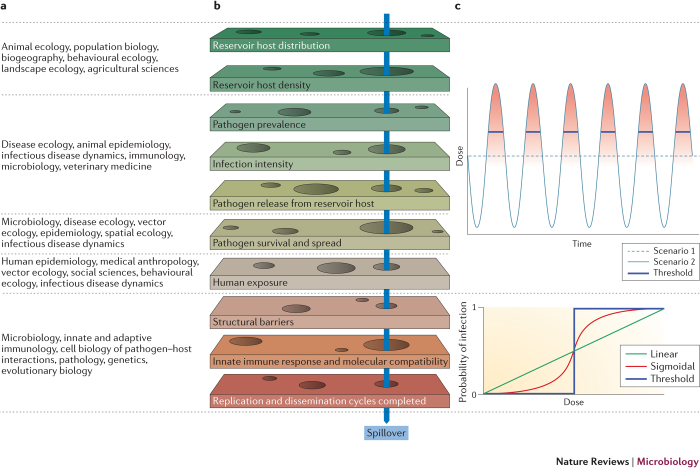
Figure 2. Barriers to spillover and dose–response relationships.
a | Determinants of spillover are being studied by researchers in many disciplines. b | A pathogen must overcome a series of barriers to transmit from one species to another. If any of these barriers is impenetrable, spillover cannot occur. Spillover of some pathogens requires that gaps (depicted as holes) in all of the barriers align within a narrow window in space and time (indicated by the blue arrow, see Supplementary information S2 (movie)). For other pathogens, protracted survival in the environment (for example, Bacillus anthracis spores109), or wide dissemination (for example, the spread of aerosolized Coxiella burnetii by wind35), may stagger the alignment of barriers to spillover. c | Top panel: hypothetical dose available over time for a given pathogen. In scenario 1 (dashed light blue line), the pathogen is excreted consistently from infected reservoir hosts. In scenario 2 (solid light blue line), the pathogen is excreted in regular but short high-intensity pulses over time. In both scenarios, the mean dose over the time interval is the same. Bottom panel: the likelihood that this dose will translate into infection depends on the functional form of the dose–response relationship. If the dose–response relationship is linear (green line), these two excretion scenarios generate the same total probability of spillover over the time interval shown. However, for nonlinear dose–response relationships, the total probability of spillover differs between scenarios. If the relationship is sigmoidal (red line), there is some probability of spillover whenever the dose exceeds zero (indicated by the intensity of the red shading in the top panel), but the total spillover probability in scenario 2 is markedly higher. In the extreme case in which the recipient host can be infected only by a dose that exceeds a sharp threshold, as suspected for Bacillus anthracis67,68,79, the pathogen in scenario 2 will spill over when the dose peaks above the threshold (blue solid line near peak), but the pathogen in scenario 1 will never spill over.
This Opinion article focuses on spillover transmission, strictly defined as the processes that enable a pathogen from a vertebrate animal to establish infection in a human. Although many recent articles have examined the fields of zoonoses or emerging pathogens2,3,10,11,12,13,14,15, a synthetic mechanistic understanding of animal-to-human transmission is lacking14,16. Much attention has been dedicated to the characterization of emerging infections3,11,12,15; for example, the high frequency of zoonoses among emerging infections3,12, their socio-economic, environmental and ecological drivers2,13,17,18, and their phylogenetic and geographical distribution3. Similarly, the phases of zoonotic emergence in the human population11,14,18, adaptation and compatibility of zoonoses in humans10,11,19, and approaches to modelling the transmission of zoonoses14,16, have also been addressed in the literature. However, a comprehensive understanding of the processes that enable a pathogen from a vertebrate animal to establish infection in a human, and how these processes are hierarchically, functionally and quantitatively linked, remains a fundamental deficit in research on zoonoses14,16. In this Opinion article, we present a mechanistic structure that integrates the determinants of spillover and the interactions among them (Fig. 1). However, we do not address broader determinants of pathogen emergence or factors that affect disease severity or onward transmission in humans.
Although many of the individual determinants of spillover are subjects of intensive study, each is usually addressed in isolation in a specialized discipline (Fig. 2). Accordingly, the better-characterized factors become the focus of public health interventions. For example, reservoir hosts or vectors are often targeted for control before the concatenation and relative influence of processes that lead to spillover are understood, which sometimes leads to inefficient or even counterproductive interventions20. In other cases, multiple mechanisms are aggregated in analyses that obscure the interactions or heterogeneities that drive risk. Although the aggregation of mechanisms may be appropriate at times, identifying discrete mechanisms and how they interact to drive spillover is essential to recognize the assumptions that are implicit in simpler models, and to clarify which processes must be modelled explicitly and which can be combined. For example, does assessment of the risk of acquiring a zoonotic infection require the measurement of the pathogen burden carried by individual reservoir hosts, or is it sufficient to estimate the cumulative abundance of a pathogen in the environment over time? This is a key question for pathogens such as Leptospira interrogans, Giardia spp., and Escherichia coli O157, and the answer may depend on modes of contact and dose–response relationships in humans (see below). Models that integrate data from experiments, the field and epidemiological studies, even if only partially parameterized, may be necessary to make such determinations.
We describe how pathogens overcome a series of barriers to pass from reservoir hosts to humans. Crucially, nonlinear interactions among the barriers create bottlenecks in the flow of a pathogen between species. Such bottlenecks provide opportunities for public health interventions that could lead to substantial reductions in the risk of spillover. Alternatively, changing environmental or social conditions can alleviate these bottlenecks, which can cause surges in spillover infections. Our framework provides the foundation for operational models that are required for quantitative evidence-based risk analysis, preparedness, surveillance and control.
Barriers to spillover
The probability of spillover is determined by the interactions among the barriers and the associated bottlenecks that might prevent cross-species transmission. Many of these interactions are nonlinear and dynamic in space and time.
Pathogen pressure. The series of processes that culminate in pathogen pressure (the amount of a pathogen that is available to humans at a given point in time and space) includes pathogen dynamics in reservoir hosts, pathogen release from reservoir hosts, and pathogen survival or dispersal outside of reservoir hosts.
Pathogen dynamics in reservoir hosts can be represented as three variables that determine the distribution and intensity of infection in time and space: the density of reservoir hosts, the prevalence of infection among reservoir hosts, and the average intensity of infection in an infected reservoir host in time and space (Supplementary information S1 (box)). Many ecological and physiological factors influence these variables in communities of reservoir animals; however, two sets of factors are dominant. The first set is the natural history of infection in hosts, which includes the duration, intensity and severity of infection and the level of shedding. Second, the movement and behaviour of hosts affect contact and the likelihood of exposure within and between species. These factors interact with the abundance, density, demographic turnover, spatial distribution and physiological state of hosts to determine the efficiency of spread21. Collectively, these processes determine how the pathogen is distributed across reservoir host populations. Such pathogen distribution can be highly variable (for example, pulses of Sin Nombre virus infections in deer mice (Peromyscus maniculatus) populations in response to climate-driven increases in population density)22, or stable (as illustrated by Mycobacterium bovis infections in populations of livestock and wildlife)23.
The mode of pathogen release from reservoir hosts determines the major routes of transmission. Pathogens may be released in host excretions, through slaughter or through an arthropod vector (Fig. 1). The probability of a pathogen being released from a reservoir host is affected by its presence and viability in relevant tissues, such as the blood for many vector-borne pathogens, tissues contacted or consumed during butchering and eating for some food-borne pathogens, and tissues through which external shedding occurs for direct or environmental routes. For example, the viral load and excretion rates in the salivary glands are key determinants for the transmission of rabies virus from carnivores, whereas viral loads in the intestinal and respiratory tracts affect the transmission of avian influenza virus from poultry24,25,26. Likewise, the release of pathogenic Leptospira spp. from animal hosts requires colonization of the renal tubules27. The excreted pathogen load depends on the quantity of leptospires that effectively colonize the tubules28, the rate of release and the urinary output of the host29. Moreover, the pathogen undergoes several changes in its lipopolysaccharide content and proteome during colonization and shedding in the urine30,31, which suggests that priming in the renal milieu is required to adapt for survival and infectivity in the external environment. The rate of pathogen release is a crucial determinant of spillover risk, and care must be taken to appropriately formulate models that represent the rate of release for each route of transmission (Box 1; Supplementary information S1 (box)).
Following the release of a pathogen from its reservoir host, the opportunity for spillover transmission is influenced by the duration of pathogen survival outside of its host, the extent of spatial dispersal through passive transport (for example, through water, on fomites or in the air), and possible pathogen reproduction or obligate developmental stages outside of the primary host (for example, Yersinia pestis, the causative agent of plague, must multiply within flea vectors before it can be transmitted to humans32). These processes can be represented as the probability that the pathogen (shed, harvested or colonized in a vector) survives and is infectious at a given point in time, and is dispersed or transported to a particular location (Supplementary information S1 (box)). Spillover of pathogens that have short survival times (for example, influenza A virus when transmitted through the respiratory route)33,34 may require close interactions between reservoir and recipient hosts. Consequently, spillover patterns in recipient hosts correspond to the prevalence patterns in reservoir hosts. By contrast, if pathogens survive for sufficient periods of time outside of their reservoir hosts, they may be dispersed beyond the home range of the host through fomites or environmental transport. In this case, the release of a pathogen from its reservoir host and human exposure to the pathogen may become disconnected in space and time. An example is the spread of aerosolized Coxiella burnetii by wind, which can lead to outbreaks of Q fever in humans that live several kilometres from the livestock reservoir hosts35.
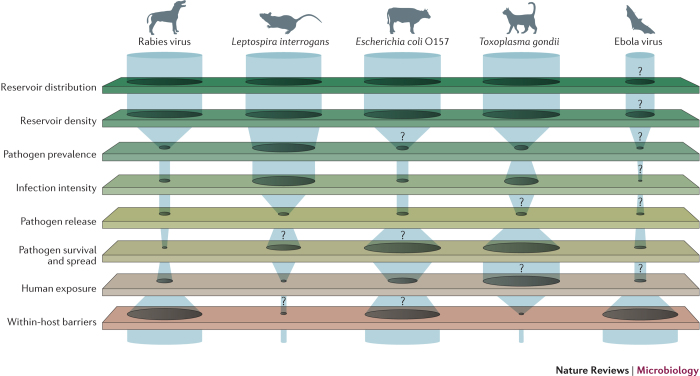
As illustrated by rabies virus, pathogenic Leptospira spp. and E. coli O157 (Fig. 3), the bottlenecks that hinder the transfer of pathogens between species depend on the ecology of the reservoir host and the pathogen, and the interactions among the determinants of spillover. For example, theprimary driver of pathogen pressure for rabies virus is the prevalence of infection in key hosts (such as domestic dogs36). Nonlinearities in rabies transmission generate a threshold effect in susceptible host density below which the pathogen cannot persist. These thresholds can be used to set vaccination targets for disease elimination37. By contrast, pathogen pressure of L. interrogans is also affected by fluctuations in reservoir host density (such as rodents29), and prevalence and shedding from infected animals29. However, if human exposure occurs through mechanisms that aggregate and disperse pathogens shed by many individuals (through accumulation in the environment, sustained survival after exiting the host38, and dispersal through rain, rivers and flood waters39), the detailed dynamics in reservoir hosts do not matter because they get integrated out by the environmental reservoir. In this scenario, spillover risk is determined by the aggregate pathogen pressure, human behaviours that determine exposure and the integrity of within-human barriers to infection. For example, when flooding mobilizes Leptospira spp. during the wet season in Brazil, human exposures can become widespread and epidemics of spillover infection can occur40. During these extreme environmental events, control efforts must focus on preventing exposure to contaminated sources (for example, by wearing protective clothing and boots41) and reducing the infectious inoculum rather than reducing the source of pathogen shedding, as the release of Leptospira spp. into the environment by animal reservoirs occurs before the extreme precipitation. Similarly, pathogen pressure of E. coli O157 is affected by the density of its cattle host population42, by variation in shedding among individuals and by prevalence in herds43. Each of these factors can be highly skewed and seasonal44,45. If spillover events are driven by contact between humans and cattle, then variation in pathogen load among animals would interact with nonlinear dose–response functions to determine spillover risk (see below). However, this individual variation matters less if human exposure occurs after human-mediated dispersal of the pathogen through irrigation, meat processing and food transportation46,47,48. In this instance, outbreaks of E. coli O157 are determined by the pathogen pressure on vegetables or in hamburger meat, potentially derived from many sources. As the dose that is required for E. coli O157 spillover is thought to be very low49,50, public health policies aim to completely eliminate pathogen pressure in food that is processed for human consumption50. To achieve this goal, interventions are focused on creating successive bottlenecks in several barriers to spillover, including decreasing cattle density, preventing faecal contamination during meat processing and increasing cooking temperatures to reduce exposure dose in ground beef43,47,51. Cumulatively, these efforts are usually successful, but high levels of shedding from cattle during summer can sometimes overwhelm interventions47.
Figure 3. Bottlenecks to spillover.
Different barriers permit or constrain the flow of pathogens from one species to another. The figure is illustrative, owing to the lack of sufficient data for more than one or two barriers for any given system. The width of the gaps in barriers represents the ease with which a pathogen can flow through the barriers and will vary depending on context. The question marks represent points at which the barriers are especially poorly understood and highlight gaps in our knowledge of some pathogens that are of global concern (for example, the lack of information on disease dynamics in reservoir hosts of Ebola virus). Many rabies virus reservoirs, such as domestic dogs, are widely distributed. The prevalence of rabies virus is generally low and the incidence of spillover closely tracks the prevalence of infection in the reservoir host. Rabies virus is almost always fatal to spillover hosts25. Interventions are usually aimed at reducing the prevalence in reservoir hosts through vaccination37. Leptospira interrogans survives in water and soil after being shed in the urine of a wide range of rodents and other reservoir hosts29. Key bottlenecks to the zoonotic spillover of this pathogen are exposure and within-host barriers. For example, during floods in Brazil, many humans that are exposed do not become infected, probably because the initial within-host barrier, the skin, is not penetrated41. However, once L. interrogans penetrates the skin (for example, through skin wounds), 1–10 leptospires may be sufficient to cause systemic infection110. Therefore, wearing protective clothing and boots is an effective control measure41. Important bottlenecks to Escherichia coli O157 spillover include heterogeneous shedding from cattle43,44 (although it is still unknown whether super-shedding is a characteristic of particular individuals or is a transient phase that occurs in most cattle42). In some contexts, exposure is an important bottleneck; for example, when the pathogen is eliminated from food through cooking. Widespread dispersal leads to uncertainties about the source of many outbreaks46,47, and weak within-human barriers enable low doses of E. coli to cause infection49,50. Humans are frequently exposed to Toxoplasma gondii carried by domestic cats and intermediate hosts, but the parasite rarely causes disease because most humans have strong within-host immunological barriers. Cats are widely and densely distributed, but the prevalence of T. gondii is low and cats shed oocysts only once in their lifetime111. However, sporulated oocysts survive in the environment for long periods of time112. Limiting exposure to oocysts may prevent spillover; however, this is challenging when it is unclear whether cats or the environment are the major sources of infection in humans111,113. Ebola virus has not been isolated from bats and the definitive reservoir bat species is unknown114; therefore, characteristics of infection in bats are unknown114,115. The pathogen is released through excretion or slaughter, then survives for up to a week, depending on the environmental conditions116. The most tractable bottlenecks for intervention may be the zoonotic exposure of humans through interaction with bats, bushmeat or the carcasses of other species97,117,118, because once exposed, the within-host barriers to Ebola virus may be extremely low119.
Exposure. The next phase of spillover — exposure — bridges the upstream processes that generate pathogen pressure and the within-host processes in the recipient that determine whether a given dose generates a spillover infection (see below). The interaction between recipient hosts and pathogen pressure determines both the dose and the route of exposure. Different behaviours of the recipient host are relevant to exposure through different routes of transmission52. Human behaviours, such as occupational interactions with reservoir host animals, the consumption of certain animal products or the use of particular environments, may increase the risk of infection53.
Exposure is often conceptualized as a simple point of contact. However, nonlinear interactions between pathogen pressure, human risk behaviour and environmental factors can lead to unexpected complexity, especially for vector-borne diseases. For example, in rats, both a high prevalence of Y. pestis and high mortality may be necessary to drive outbreaks of bubonic plague in humans. Widespread exposure of humans through flea bites occurs only after a decrease in the abundance of rats, which are the primary hosts of Y. pestis in peridomestic settings54. Indeed, historically, high rat mortality ('rat-fall') was an indication of an imminent human plague epidemic32. Thus, killing rodents in response to cases of bubonic plague in humans could inadvertently increase the severity of the epidemic54. Conversely, and controversially, zooprophylaxis, which involves diverting vector bites from humans by increasing the local population density of another animal host, may decrease the risk of human exposure55. For example, the presence of chickens and dogs in rural areas of Argentina decreased the rate at which Triatoma infestans transmitted Trypanosoma cruzi, the causative agent of Chagas disease, to humans56. However, increasing the population density of reservoir hosts may also affect vector survival, vector abundance and pathogen prevalence in reservoir hosts, which, in turn, increases pathogen pressure and offsets reductions in human–vector contact rates56,57. These complexities highlight the need to understand the mechanisms that contribute to particular routes of spillover.
All of the factors that precede human exposure, mediated by human behaviour and environmental factors (Fig. 1), cumulate in the dose to which a host is exposed at a given location and time (the integral of the pathogen pressure in space and time to which the host has been exposed (Supplementary information S1 (box)).
Probability of infection. Following cross-species exposure of a recipient host, the within-host barriers and their interactions with the strain of pathogen determine the functional relationship between the pathogen dose and the likelihood that an infection will establish. Within-host barriers to infection vary widely and depend on the specific combinations of pathogen, host species and individual receptivity58. Physical barriers include the skin, mucous membranes, mucus, stomach acid or the absence of functional receptors that enable the pathogen to enter its target cells or tissues10. Interferon-induced and other innate immune responses may be triggered after the initial infection of a cell, resulting in protective mechanisms such as apoptosis or the induction of interferon-induced resistance in surrounding cells59. In addition, interfering defensive proteins in the host cell cytoplasm may block the replication of intracellular pathogens. In other cases, cells lack functional host factors that are required for the replication of the pathogen60,61. Even when pathogens can replicate within cells, several barriers can prevent their transmission to other cells62,63 and thus the establishment of an infection. For example, avian influenza virus must pass through a series of within-host barriers to infect a human, including mucins in respiratory tract excretions, specific receptor molecules that constrain virus entry into cells and have different distributions in the respiratory tracts of different host species, suboptimal viral polymerase that restricts the ability of the virus to replicate in cells of the human respiratory tract, viral neuraminidase that is inefficient in its role in the release of influenza viruses from infected cells, and innate immune responses that are initiated early and that block infection in both infected and neighbouring cells63,64.
From an epidemiological perspective, these within-host interactions between zoonotic pathogens and hosts can be encapsulated by the functional relationship between pathogen dose and the probability of an infection. Although there is much to learn about dose–response relationships, they are expected to be nonlinear as, at minimum, they must saturate at high doses because the probability of infection cannot exceed one65. This nonlinearity imposes a filter on the dynamics of pathogen pressure and exposure (Fig. 2c). If the dose–response relationship is highly nonlinear, such that small changes in dose lead to large changes in the probability of an infection, then variation in any of the upstream factors that culminate in an exposure dose (including released dose, pathogen survival and human behaviour) may have disproportionate effects on the probability of spillover. Such effects could generate opportunities for targeted control measures. Moreover, nonlinear dose–response relationships may imply that infrequent high-intensity exposures are more likely to cause spillover infections than continuous low-intensity excretion. This phenomenon has been reported for occupational exposure to Bacillus anthracis aerosols; tannery workers who were exposed to infrequent high doses of B. anthracis spores in imported goat hair were more likely to die of anthrax than those who were exposed to frequent low doses of B. anthracis spores66,67,68. Conversely, if doses are far below the inflection point on the dose–response curve (Fig. 2c), then the system may be insensitive to changes in dose. If the dose–response function is close to linear, the total exposure dose over time is equal and host responses do not change as a consequence of early exposures, then longer-term exposure to a low but constant dose may generate the same probability of infection as intermittent high-intensity exposures (Fig. 2c).
The genetic, immunological and physiological state of the host also can modulate the dose–response relationship. Immunosuppression (for example, due to AIDS, immunosuppressive drugs, co-infections or malnutrition) increases gaps in within-host barriers, which shifts dose–response curves and increases susceptibility69,70. For example, in immunosuppressed hosts, the decreased number or activity of lymphocytes can reduce the dose that is required to establish an infection with the widespread pathogen Toxoplasma gondii, or cause the loss of control of T. gondii infections that are usually kept in check by sustained immune pressure71 (Fig. 3). Seasonality in human immune function (for example, enhanced baseline inflammation and altered cellular composition of the immune system in winter compared with summer) may also alter the permeability of within-host barriers by altering the magnitude and speed of immune responses72. Finally, the probability and severity of infection at a given dose are shaped by host genetics73; triathletes with a particular gene polymorphism were at increased risk of leptospirosis after swallowing lake water compared with athletes who lacked this polymorphism74.
Many of the interactions at the crossroads of exposure, inoculum dose and host response are poorly understood. Therefore, very little is known about the interactions between dose, timing of exposure and probability of infection. The current dose–response paradigm is based on discrete transient exposures, but the effects of protracted or cumulative exposure to environmental pathogens (for example, to low concentrations of Leptospira spp. in floodwater) are unclear75. Repeated low-dose exposure can increase host immunity to infection (for example, as postulated for poultry handlers who are exposed to avian influenza76, dairy farmers who are exposed to E. coli O157 (Ref. 77) and mice that are exposed to continuous infections of parasites78). However, increases in immunity are not always observed; for example, such effects on immunity were not observed in tannery workers who were exposed to B. anthracis67,68,79. Moreover, it may be difficult to differentiate between a cumulative dose effect and the increasing opportunity to initiate an infection with each additional low-dose exposure (if each infectious unit has a probability of causing an infection that is above zero)20,80.
Once a pathogen has penetrated the within-host barriers to replicate and disseminate in the new host, the outcome of the infection may range from subclinical elimination of the microorganism to the death of the new host, and from dead-end spillover infection to sustained human-to-human transmission. For many important zoonotic pathogens, such as HIV or Zika virus, the transmission that drives the current public health crisis is human-to-human81,82 and the events that led to spillover are long past. Although understanding disease severity and onward transmission is essential for understanding the consequences of emerging infectious diseases, these processes are beyond the scope of this article. Our current knowledge of the biological features of pathogens and characteristics of host–pathogen interactions that determine these outcomes are described elsewhere (for example, see Refs 83,84).
Box 1: The mathematics of spillover.
The opportunities for cross-species transmission are influenced by processes that occur at scales from molecules to landscapes (Fig. 1). These processes are subjects of intense study, and their characterization is complicated by their variability in space and time, nonlinear responses and interactions with outside factors. Consequently, it is impossible to integrate all of the determinants of spillover transmission — or to assess the effects of gaps in our knowledge about these determinants — without appropriate tools, such as mathematical and computational models107.
In Supplementary information S1 (box), we present a general mathematical model of the spillover process, which provides a template for integrating our knowledge of processes for specific disease systems. This model framework essentially translates Fig. 1 into mathematical expressions. It allows for variation in space and time, and uses different formulations for transmission through pathogen excretion, slaughter or arthropod vectors.
The mathematical model reflects the modular nature of the spillover process, as emphasized in the main text, while highlighting dependencies among factors in ways such as the following:
• Factors that are linked to disease ecology of the reservoir host and the mode of pathogen release determine the amount of pathogen released to the environment or vector.
• Pathogen survival and transport outside of the animal host, which give rise to pathogen pressure at a particular place and time, are modelled with simple probability kernels.
• Human risk behaviours determine how this pathogen pressure translates to exposure dose.
• The probability of infection for a given dose and route of exposure is encapsulated in the dose–response relationship (Fig. 2c).
Mathematically, the focal point of this process is the dose to which the recipient host is exposed. All upstream factors come together, with appropriate functional dependencies, to shape this dose. To a reasonable approximation, which is consistent with current practice in quantitative microbial risk assessment108, the consequent risk of infection can be modelled independently through the dose–response relationship.
Assessing zoonotic risk
When gaps in barriers to spillover are highly dynamic in time and space, they may vary asynchronously, so that the alignment of gaps in all barriers may be fleeting and spillover may seem random (Supplementary information S2 (movie)). Research methods that group multiple barriers or integrate data over space and time may not capture these dynamics. For example, ecological niche models are often used to study zoonotic risk by assessing the distribution of reservoir hosts or vectors85, but this approach overlooks variation in downstream barriers that might drive risk. Alternatively, niche models that are based on the documented occurrence of spillover may capture the accumulated distribution of all conditions that enabled barriers to be breached over time (Fig. 1), but they cannot isolate the precise barriers that affect spillover risk (for example, see Ref. 86). Therefore, niche models tend to overestimate the spatial range of spillover risk and do not readily enable extrapolation to novel conditions87. Examples of this include Hendra virus and Marburg virus, which can be excreted in discrete temporal and spatial pulses from their bat reservoir hosts20,88,89. However, for spillover, shedding must align with environmental and bat population conditions that generate levels of pathogen pressure that are sufficient to produce an infectious dose (Fig. 2), and with exposure behaviours and susceptibility of the recipient hosts. As some of these conditions vary among seasons and years, the pattern of outbreaks in livestock or humans has high spatial and temporal variability20,89. However, as niche models often summarize risk across large areas and long durations, they overlook important heterogeneities and they lack the specificity that is required for public health intervention. Although niche models can help to identify regional-to-continental concentrations of risk90,91, risk assessments that are more quantitative and more precise with regard to space, time and which barriers they address are needed to guide concrete action.
Epidemiological investigations of spillover also need to account for conditions that are highly dynamic in space and time. If the alignment of gaps in all barriers is fleeting, delayed diagnoses or inconsistent case detection may delay outbreak investigations until the conditions that enabled spillover have changed. Similarly, investigations are sometimes triggered once the case count becomes high. These challenges differ among pathogens with different values of R0 (the basic reproductive number or expected number of secondary infections caused by a typical infected individual in a susceptible population). For supercritical pathogens with R0 >1, which can cause major epidemics through sustained transmission in human populations (for example, Ebola virus, Zika virus and the pandemic strain of severe acute respiratory syndrome coronavirus (SARS-CoV)4,81,92), spillover becomes challenging to study because a given human case is likely to be far removed in time or space from the spillover event that triggered an outbreak. Subcritical pathogens with 0 <R0 <1, which cause self-limiting outbreaks or 'stuttering chains' in human populations (for example, monkeypox or avian influenza viruses93,94), raise distinct challenges because any given individual could have been infected by either an animal or a human source16. It is easiest to study the spillover of pathogens with R0 = 0 that are not transmitted between humans (for example, rabies virus or West Nile virus25,95), in which every case is an instance of spillover. The 2014–2015 Ebola virus epidemic in West Africa is a prime example whereby delayed response and investigation prevented researchers from reconstructing the conditions that initiated the human epidemic of a supercritical pathogen96,97. Ebola virus infection is an extreme example of spillover infection that only occurs during the rare alignment of gaps in barriers, and, accordingly, the precise determinants of risk are poorly understood (Fig. 3). By contrast, for other zoonoses, such as trypanosomiasis in some parts of Africa, incidence is high because the pathogen flows through consistently wide gaps in barriers to infection (for example, common exposure to infected animal hosts and tsetse fly vectors, and low resistance in humans due to the ability of trypanosomes to neutralize or avoid human innate immune activity98,99). In all scenarios, irrespective of the frequency with which gaps align, the concept of hierarchical barriers can be used to organize and quantify the conditions that enable spillover.
The influence of particular barriers may vary in space and time, and this variation — coupled with data on realized spillover events — can help elucidate factors that shape infection risk, even in the absence of information on other barriers. In the westernmost province of the Democratic Republic of Congo, the observed lack of monkeypox spillover, despite high seroprevalence in the suspected reservoir hosts (Heliosciurus spp. and Funisciuris spp.), was attributed to cultural norms that forbade the consumption of small rodents100. The inconsistency between ecological data that suggested high pathogen pressure and epidemiological data that indicated a lack of spillover, focused attention on human behaviours that affect the probability of exposure. Research approaches that integrate data on multiple barriers are more likely to discern such behavioural effects.
Broad-scale discovery of novel microorganisms has the potential to characterize the pool of possible zoonotic pathogens and provide valuable baseline information101,102. However, each of the ∼63,000 species of mammals, birds, reptiles, amphibians and fish103 contains a multitude of infectious viruses, bacteria and parasites101,102,104,105,106. Although each of these microorganisms and parasites can be viewed as a potential pathogen, the vast majority may not cause disease in their natural hosts, and the extent to which they infect or cause pathology in other species, including humans, is unknown7,9,10. Therefore, discovery alone cannot address the potential risk of spillover. The translation of new discoveries of microorganisms into guidance for public health practitioners requires the identification of the barriers to microbial infection of humans, the conditions that facilitate the breaching of these barriers, and, therefore, the microbiological and environmental contexts that pose the greatest risk to human populations. For the foreseeable future, the greatest practical contribution of pathogen discovery and sequence characterization to the epidemiology of emerging pathogens is likely to be in the rapid post hoc identification of novel pathogens after spillover.
Outlook
The framework presented in this Opinion article highlights that an important frontier in research on zoonotic spillover is to understand the functional and quantitative links among the determinants of spillover. To our knowledge, all of the processes that are necessary to achieve spillover have not been connected, compared and quantified for any single zoonotic pathogen. We address this gap, in part, by introducing a conceptual and quantitative model that can be used to integrate existing data, identify high-priority data gaps, investigate conditions that widen or align gaps in barriers to spillover, and identify the best gaps on which to focus intervention efforts. We suggest that future research focuses on developing case studies that contribute to fully quantifying the determinants of spillover and their linkages, with the goal of making operational contributions to risk assessment. We provide a mathematical framework that formalizes the ideas presented here to guide the formulation of mechanistic spillover models for particular zoonotic pathogens (Box 1; Supplementary information S1 (box)). We anticipate that this synthetic framework will provide a foundation for cross-scale data integration, transdisciplinary investigation, and a new body of theory on spillover that is necessary for risk assessment and public health planning.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
A mathematical representation of spillover (PDF 110 kb)
Spatiotemporal dynamics of spillover. Gaps in barriers to spillover may be highly dynamic in time and space meaning that the alignment of gaps in all barriers may be rare and brief. (GIF 2542 kb)
Acknowledgements
The authors thank J. Wood and E. Fleishman for helpful comments and conversations. R.K.P. and H.M. are supported by the Commonwealth of Australia, the State of New South Wales and the State of Queensland under the National Hendra Virus Research Program, awarded through the Rural Industries Research and Development Corporation. R.K.P. is supported by the US National Institutes of General Medical Sciences IDeA Program (grants P20GM103474 and P30GM110732), P. Thye, the Morris Animal Foundation, Montana University System Research Initiative (grant 51040-MUSRI2015-03), a Defense Advanced Research Projects Agency (DARPA) Young Faculty Award and the US Department of Defense Strategic Environmental Research and Development Program (SERDP; grant RC-2633). J.O.L.-S. is supported by the US National Science Foundation (NSF; grants OCE-1335657 and DEB-1557022) and the US Department of Defense SERDP (grant RC-2635). J.O.L.-S., A.L.G. and P.J.H. are supported by the RAPIDD program of the Science & Technology Directorate of the Department of Homeland Security, the Fogarty International Center (part of the US National Institutes of Health), and by IDEAS (Infectious Disease Evolution Across Scales), which is a Research Coordination Network (DEB-1354890) funded by the US National Science Foundation.
Biographies
Raina K. Plowright is Assistant Professor of Epidemiology at Montana State University, Bozeman, USA. She received her veterinary degree from the University of Sydney, Australia, and then travelled to the USA as an Australian Fulbright and Centennial Scholar to do her Ph.D. in ecology and M.Sc. in epidemiology at the University of California, Davis, USA. She was a David H. Smith Conservation Research Fellow at the Center for Infectious Disease Dynamics at Pennsylvania State University, USA. Her group at Montana State University studies pathogens that spill over from animals to people, the dynamics of zoonotic pathogens in wildlife populations, and pathogens that threaten wildlife conservation.
Colin R. Parrish is the John M. Olin Professor of Virology in the Baker Institute for Animal Health at the College of Veterinary Medicine, Cornell University, Ithaca, New York, USA. His research focuses on the study of viruses, virus structures and the evolution of new viral host ranges. His studies also examine the general basis of viral emergence, in particular the risk factors that are associated with the origins of new viruses in humans or other animals. For example, he studies canine parvovirus and canine influenza viruses emerging to cause epidemics in dogs.
Hamish McCallum is a professor in the Griffith School of Environment and Environmental Futures Institute at Griffith University, Queensland, Australia. After his B.Sc. at Monash University, Melbourne, Victoria, Australia, he completed his Ph.D. under the supervision of Roy Anderson at Imperial College London, UK. His research primarily focuses on the ecology of infectious diseases in wildlife using quantitative approaches. The systems he works on include Tasmanian devil facial tumour disease, the amphibian chytrid fungus, Hendra virus in pteropid bats and chlamydial disease in koalas.
Peter J. Hudson is the Willaman Professor of Biology and Director of The Huck Institutes of Life Sciences at Pennsylvania State University, USA. He investigates the dynamics of infections in free-living animal populations, spillover between host species, and patterns of invasion and consequences for wildlife populations. He received his doctorate from the University of Oxford, UK, and he was elected a fellow of the Royal Society in 2008.
Albert I. Ko is Professor of Epidemiology and Department Chair of Epidemiology of Microbial Diseases at the Yale School of Public Health, New Haven, Connecticut, USA. His research centres on the health problems that have emerged as a consequence of rapid urbanization and social inequity. He coordinates a research and training programme on urban slum health in Brazil and is conducting prospective community-based studies on rat-borne leptospirosis, dengue, meningitis and respiratory infections. More recently, his team has mobilized the public health research capacity at their site in the city of Salvador, Brazil to investigate the ongoing outbreak of Zika virus and resulting microcephaly.
Andrea L. Graham is Associate Professor of Ecology and Evolutionary Biology at Princeton University, New Jersey, USA. She earned her A.B. in biology and sculpture from Mount Holyoke College, South Hadley, Massachusetts, USA, and her Ph.D. in ecology and evolutionary biology from Cornell University, Ithaca, New York, USA, before completing her postdoctoral training at the University of Edinburgh, UK. She then held 2 independent research fellowships at Edinburgh and moved to Princeton University in 2011. Her research interests centre on the causes and consequences of immunological heterogeneity.
James O. Lloyd-Smith is Professor in the Departments of Ecology and Evolutionary Biology, and Biomathematics at University of California, Los Angeles, USA. His research programme explores the ecological and evolutionary dynamics of infectious diseases in animal and human populations, with emphasis on the emergence of zoonotic pathogens and drug-resistant strains. His group combines mathematical models, statistical analysis, and laboratory, clinical and field studies to learn about diseases such as monkeypox, leptospirosis and influenza. He received his Ph.D. from the University of California, Berkeley, USA, where he studied heterogeneity in disease transmission dynamics, and he carried out his postdoctoral studies at Pennsylvania State University, USA.
PowerPoint slides
Competing interests
The authors declare no competing financial interests.
References
- 1.Christou L. The global burden of bacterial and viral zoonotic infections. Clin. Microbiol. Infect. 2011;17:326–330. doi: 10.1111/j.1469-0691.2010.03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briand S, et al. The international Ebola emergency. N. Engl. J. Med. 2014;371:1180–1183. doi: 10.1056/NEJMp1409858. [DOI] [PubMed] [Google Scholar]
- 5.Smith GJ, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 6.Fevre EM, Wissmann BV, Welburn SC, Lutumba P. The burden of human African trypanosomiasis. PLoS Negl. Trop. Dis. 2008;2:e333. doi: 10.1371/journal.pntd.0000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, Segre JA. The skin microbiome. Nat. Rev. Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 10.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolhouse ME, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morse SS. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd-Smith JO, et al. Epidemic dynamics at the human–animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CK, et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 2015;5:14830. doi: 10.1038/srep14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd-Smith JO, Funk S, McLean AR, Riley S, Wood JL. Nine challenges in modelling the emergence of novel pathogens. Epidemics. 2015;10:35–39. doi: 10.1016/j.epidem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gortazar C, et al. Crossing the interspecies barrier: opening the door to zoonotic pathogens. PLoS Pathog. 2014;10:e1004129. doi: 10.1371/journal.ppat.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pepin KM, Lass S, Pulliam JR, Read AF, Lloyd-Smith JO. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat. Rev. Microbiol. 2010;8:802–813. doi: 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plowright RK, et al. Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. B Biol. Sci. 2015;282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson PJ, Rizzoli AR, Grenfell BT, Heesterbeek H, Dobson AP. The Ecology of Wildlife Diseases. 2002. [Google Scholar]
- 22.Hjelle B, Glass GE. Outbreak of hantavirus infection in the Four Corners region of the United States in the wake of the 1997–1998 El Nino—Southern Oscillation. J. Infect. Dis. 2000;181:1569–1573. doi: 10.1086/315467. [DOI] [PubMed] [Google Scholar]
- 23.Thoen CO, Steele JH, Kaneene JB. Zoonotic Tuberculosis: Mycobacterium bovis and Other Pathogenic Mycobacteria. 2014. [Google Scholar]
- 24.Ducatez M, Webster R, Webby R. Animal influenza epidemiology. Vaccine. 2008;26:D67–D69. doi: 10.1016/j.vaccine.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupprecht CE, Hanlon CA, Hemachudha T. Rabies re-examined. Lancet Infect. Dis. 2002;2:327–343. doi: 10.1016/s1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- 26.Webster R. Viral Zoonoses and Food of Animal Origin. 1997. pp. 105–113. [Google Scholar]
- 27.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa F, et al. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Negl. Trop. Dis. 2014;8:e3338. doi: 10.1371/journal.pntd.0003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa F, et al. Patterns in Leptospira shedding in Norway rats (Rattus norvegicus) from Brazilian slum communities at high risk of disease transmission. PLoS Negl. Trop. Dis. 2015;9:e0003819. doi: 10.1371/journal.pntd.0003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monahan AM, Callanan JJ, Nally JE. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 2008;76:4952–4958. doi: 10.1128/IAI.00511-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nally JE, Chow E, Fishbein MC, Blanco DR, Lovett MA. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect. Immun. 2005;73:3251–3260. doi: 10.1128/IAI.73.6.3251-3260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smego R, Frean J, Koornhof H. Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 1999;18:1–15. doi: 10.1007/s100960050219. [DOI] [PubMed] [Google Scholar]
- 33.Weber TP, Stilianakis NI. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J. Infect. 2008;57:361–373. doi: 10.1016/j.jinf.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koopmans M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 35.Tissot-Dupont H, Amadei M-A, Nezri M, Raoult D. Wind in November, Q fever in December. Emerg. Infect. Dis. 2004;10:1264. doi: 10.3201/eid1007.030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hampson K, et al. Synchronous cycles of domestic dog rabies in sub-Saharan Africa and the impact of control efforts. Proc. Natl Acad. Sci. USA. 2007;104:7717–7722. doi: 10.1073/pnas.0609122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brochier B, et al. Large-scale eradication of rabies using recombinant vaccinia–rabies vaccine. Nature. 1991;354:520–522. doi: 10.1038/354520a0. [DOI] [PubMed] [Google Scholar]
- 38.Andre-Fontaine G, Aviat F, Thorin C. Waterborne leptospirosis: survival and preservation of the virulence of pathogenic Leptospira spp. in fresh water. Curr. Microbiol. 2015;71:136–142. doi: 10.1007/s00284-015-0836-4. [DOI] [PubMed] [Google Scholar]
- 39.Lau CL, Smythe LD, Craig SB, Weinstein P. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 2010;104:631–638. doi: 10.1016/j.trstmh.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Reis RB, et al. Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl. Trop. Dis. 2008;2:e228. doi: 10.1371/journal.pntd.0000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phraisuwan P, et al. Leptospirosis: skin wounds and control strategies, Thailand, 1999. Emerg. Infect. Dis. 2002;8:1455–1459. doi: 10.3201/eid0812.020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer SE, Besser TE, Cobbold RN, French NP. 'Super'or just 'above average'? Supershedders and the transmission of Escherichia coli O157: H7 among feedlot cattle. J. R. Soc. Interface. 2015;12:0446. doi: 10.1098/rsif.2015.0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl Acad. Sci. USA. 2006;103:547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock D, Besser T, Rice D, Herriott D, Tarr P. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besser TE, Davis MA, Walk ST. Population Genetics of Bacteria: A Tribute to Thomas S. Whittam. 2011. pp. 303–324. [Google Scholar]
- 46.Gerba CP, Smith JE. Sources of pathogenic microorganisms and their fate during land application of wastes. J. Environ. Qual. 2005;34:42–48. [PubMed] [Google Scholar]
- 47.Elder RO, et al. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl Acad. Sci. USA. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pennington H. Escherichia coli O157. Lancet. 2010;376:1428–1435. doi: 10.1016/S0140-6736(10)60963-4. [DOI] [PubMed] [Google Scholar]
- 49.Teunis P, Ogden I, Strachan N. Hierarchical dose response of E. coli O157: H7 from human outbreaks incorporating heterogeneity in exposure. Epidemiol. Infect. 2008;136:761–770. doi: 10.1017/S0950268807008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuttle J, et al. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 1999;122:185–192. doi: 10.1017/s0950268898001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cobbold RN, et al. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157: H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 2007;73:1563–1568. doi: 10.1128/AEM.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cascio A, Bosilkovski M, Rodriguez-Morales A, Pappas G. The socio-ecology of zoonotic infections. Clin. Microbiol. Infect. 2011;17:336–342. doi: 10.1111/j.1469-0691.2010.03451.x. [DOI] [PubMed] [Google Scholar]
- 53.Macpherson CN. Human behaviour and the epidemiology of parasitic zoonoses. Int. J. Parasitol. 2005;35:1319–1331. doi: 10.1016/j.ijpara.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Keeling MJ, Gilligan CA. Metapopulation dynamics of bubonic plague. Nature. 2000;407:903–906. doi: 10.1038/35038073. [DOI] [PubMed] [Google Scholar]
- 55.Hess A, Hayes RO. Relative potentials of domestic animals for zooprophylaxis against mosquito vectors of encephalitis. Am. J. Trop. Med. Hyg. 1970;19:327–334. doi: 10.4269/ajtmh.1970.19.327. [DOI] [PubMed] [Google Scholar]
- 56.Gürtler RE, et al. Domestic animal hosts strongly influence human-feeding rates of the Chagas disease vector Triatoma infestans in Argentina. PLoS Negl. Trop. Dis. 2014;8:e2894. doi: 10.1371/journal.pntd.0002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kilpatrick AM, Randolph SE. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet. 2012;380:1946–1955. doi: 10.1016/S0140-6736(12)61151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmid-Hempel P. Variation in immune defence as a question of evolutionary ecology. Proc. Biol. Sci. 2003;270:357–366. doi: 10.1098/rspb.2002.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 60.Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Air GM, Laver WG. The neuraminidase of influenza virus. Proteins. 1989;6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 63.Kuiken T, et al. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 64.Lipsitch M, et al. Viral factors in influenza pandemic risk assessment. eLife. 2016;5:e18491. doi: 10.7554/eLife.18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmid-Hempel P, Frank SA. Pathogenesis, virulence, and infective dose. PLoS Pathog. 2007;3:e147. doi: 10.1371/journal.ppat.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brachman PS, Fekety FR. Industrial anthrax. Ann. NY Acad. Sci. 1958;70:574–584. doi: 10.1111/j.1749-6632.1958.tb35413.x. [DOI] [PubMed] [Google Scholar]
- 67.Brachman PS, Kaufman A, Dalldorf FG. Industrial inhalation anthrax. Bacteriol. Rev. 1966;30:646. doi: 10.1128/br.30.3.646-659.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coleman ME, Thran B, Morse SS, Hugh-Jones M, Massulik S. Inhalation anthrax: dose response and risk analysis. Biosecur. Bioterror. 2008;6:147–160. doi: 10.1089/bsp.2007.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bollaerts K, et al. Human salmonellosis: estimation of dose–illness from outbreak data. Risk Anal. 2008;28:427–440. doi: 10.1111/j.1539-6924.2008.01038.x. [DOI] [PubMed] [Google Scholar]
- 70.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greene CE. Infectious Diseases of the Dog and Cat. 2013. [Google Scholar]
- 72.Dopico XC, et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 2015;6:7000. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gingles NA, et al. Role of genetic resistance in invasive pneumococcal infection: identification and study of susceptibility and resistance in inbred mouse strains. Infect. Immun. 2001;69:426–434. doi: 10.1128/IAI.69.1.426-434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lingappa J, et al. HLA-DQ6 and ingestion of contaminated water: possible gene–environment interaction in an outbreak of leptospirosis. Genes Immun. 2004;5:197–202. doi: 10.1038/sj.gene.6364058. [DOI] [PubMed] [Google Scholar]
- 75.Pujol JM, Eisenberg JE, Haas CN, Koopman JS. The effect of ongoing exposure dynamics in dose response relationships. PLoS Comput. Biol. 2009;5:e1000399. doi: 10.1371/journal.pcbi.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang W, Shaman J. Does exposure to poultry and wild fowl confer immunity to H5N1? Chin. Med. J. 2014;127:3335. [PMC free article] [PubMed] [Google Scholar]
- 77.Reymond D, et al. Neutralizing antibodies to Escherichia coli vero cytotoxin 1 and antibodies to O157 lipopolysaccharide in healthy farm family members and urban residents. J. Clin. Microbiol. 1996;34:2053–2057. doi: 10.1128/jcm.34.9.2053-2057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scott M. High transmission rates restore expression of genetically determined susceptibility of mice to nematode infections. Parasitology. 2006;132:669–679. doi: 10.1017/S0031182005009583. [DOI] [PubMed] [Google Scholar]
- 79.Cohen ML, Whalen T. Implications of low level human exposure to respirable B. anthracis. Appl. Biosafety. 2007;12:109. [Google Scholar]
- 80.French N, Kelly L, Jones R, Clancy D. Dose–response relationships for foot and mouth disease in cattle and sheep. Epidemiol. Infect. 2002;128:325–332. doi: 10.1017/s0950268801006446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Faria NR, et al. Zika virus in the Americas: early epidemiological and genetic findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 83.Geoghegan JL, Senior AM, Di Giallonardo F, Holmes EC. Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl Acad. Sci. USA. 2016;113:4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Casadevall A, Pirofski L. Host–pathogen interactions: the attributes of virulence. J. Infect. Dis. 2001;184:337–344. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- 85.Miller RH, et al. Ecological niche modeling to estimate the distribution of Japanese encephalitis virus in Asia. PLoS Negl. Trop. Dis. 2012;6:e1678. doi: 10.1371/journal.pntd.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levine RS, et al. Ecological niche and geographic distribution of human monkeypox in Africa. PLoS ONE. 2007;2:e176. doi: 10.1371/journal.pone.0000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kearney M, Simpson SJ, Raubenheimer D, Helmuth B. Modelling the ecological niche from functional traits. Phil. Trans. R. Soc. B Biol. Sci. 2010;365:3469–3483. doi: 10.1098/rstb.2010.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Plowright RK, et al. Transmission or within-host dynamics driving pulses of zoonotic viruses in reservoir-host populations. PLoS Negl. Trop. Dis. 2016;10:e0004796. doi: 10.1371/journal.pntd.0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amman BR, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pigott DM, et al. Mapping the zoonotic niche of Marburg virus disease in Africa. Trans. R. Soc. Trop. Med. Hyg. 2015;109:366–378. doi: 10.1093/trstmh/trv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brierley L, Vonhof M, Olival K, Daszak P, Jones K. Quantifying global drivers of zoonotic bat viruses: a process-based perspective. Am. Nat. 2016;187:E53–E64. doi: 10.1086/684391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 93.Hutin Y, et al. Outbreak of human monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg. Infect. Dis. 2001;7:434. doi: 10.3201/eid0703.010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Q, et al. Epidemiology of human infections with avian influenza A (H7N9) virus in China. N. Engl. J. Med. 2014;370:520–532. doi: 10.1056/NEJMoa1304617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayes EB, et al. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baize S, et al. Emergence of Zaire Ebola virus disease in Guinea — preliminary report. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 97.Saéz AM, et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol. Med. 2015;7:17–23. doi: 10.15252/emmm.201404792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kieft R, et al. Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc. Natl Acad. Sci. USA. 2010;107:16137–16141. doi: 10.1073/pnas.1007074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simarro PP, et al. Estimating and mapping the population at risk of sleeping sickness. PLoS Negl. Trop. Dis. 2012;6:e1859. doi: 10.1371/journal.pntd.0001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jezek Z, Fenner F, Melnick JL. Monographs in Virology. 1988. pp. 119–121. [Google Scholar]
- 101.Anthony SJ, et al. A strategy to estimate unknown viral diversity in mammals. mBio. 2013;4:e00598-13. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Temmam S, Davoust B, Berenger J-M, Raoult D, Desnues C. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 2014;15:10377–10397. doi: 10.3390/ijms150610377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoffmann M, et al. The impact of conservation on the status of the world's vertebrates. Science. 2010;330:1503–1509. doi: 10.1126/science.1194442. [DOI] [PubMed] [Google Scholar]
- 104.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Turnbaugh PJ, et al. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl Acad. Sci. USA. 2008;105:11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heesterbeek H, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347:aaa4339. doi: 10.1126/science.aaa4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haas CN, Rose JB, Gerba CP. Quantitative Microbial Risk Assessment. 2014. [Google Scholar]
- 109.Mitscherlich E, Marth EH. Microbial Survival in the Environment: Bacteria and Rickettsiae Important in Human and Animal Health. 2012. [Google Scholar]
- 110.Silva, É F, et al. Characterization of virulence of Leptospira isolates in a hamster model. Vaccine. 2008;26:3892–3896. doi: 10.1016/j.vaccine.2008.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elmore SA, et al. Toxoplasma gondii: epidemiology, feline clinical aspects, and prevention. Trends Parasitol. 2010;26:190–196. doi: 10.1016/j.pt.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 112.Dubey J. Toxoplasma gondii oocyst survival under defined temperatures. J. Parasitol. 1998;84:862–865. [PubMed] [Google Scholar]
- 113.Jones J, Dubey J. Waterborne toxoplasmosis — recent developments. Exp. Parasitol. 2010;124:10–25. doi: 10.1016/j.exppara.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 114.Leroy EM, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 115.Pourrut X, et al. Spatial and temporal patterns of Zaire ebolavirus antibody prevalence in the possible reservoir bat species. J. Infect. Dis. 2007;196:S176–S183. doi: 10.1086/520541. [DOI] [PubMed] [Google Scholar]
- 116.Prescott J, et al. Postmortem stability of Ebola virus. Emerg. Infect. Dis. 2015;21:856. doi: 10.3201/eid2105.150041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leroy EM, et al. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 2009;9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- 118.Leroy EM, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 119.Judson S, Prescott J, Munster V. Understanding Ebola virus transmission. Viruses. 2015;7:511–521. doi: 10.3390/v7020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A mathematical representation of spillover (PDF 110 kb)
Spatiotemporal dynamics of spillover. Gaps in barriers to spillover may be highly dynamic in time and space meaning that the alignment of gaps in all barriers may be rare and brief. (GIF 2542 kb)