Abstract
Few infectious diseases are entirely human-specific: most human pathogens also circulate in animals, or else originated in non-human hosts. Influenza, plague, and trypanosomiasis are classic examples of zoonoses, or infections that transmit from animals to humans. The multi-host ecology of zoonoses leads to complex dynamics, and analytical tools such as mathematical modeling are vital to the development of effective control policies and research agendas. Much attention has focused on modeling pathogens with simpler life cycles and immediate global urgency, such as influenza and SARS, but vector-transmitted, chronic, and protozoan infections have been neglected, as have crucial processes such as cross-species transmission. Progress in understanding and combating zoonoses requires a new generation of models that addresses a broader set of pathogen life histories and integrates across host species and scientific disciplines.
A recent survey of all recognized human pathogens revealed that over half are zoonotic (1, 2), and nearly all of the most important human pathogens are either zoonotic or originated as zoonoses before adapting to humans (3). The three most devastating pandemics in human history, the Black Death, Spanish influenza, and HIV/AIDS, were caused by zoonoses (4), as were 60–76% of recent emerging infectious disease events (2, 5). Underlying these patterns are specific public health challenges arising from the complex multi-host ecology of zoonotic infections (6, 7), and accelerating environmental and anthropogenic changes that are altering the rates and nature of contact between human and animal populations (8–10). Following a series of recent outbreaks (e.g., avian and swine influenza, West Nile virus and SARS), a rising sense of urgency has stimulated a broad increase in research on zoonoses, ranging from dissection of the molecular determinants of host specificity (11) to viral prospecting in African rain forests (12). Such endeavors have produced important insights into underlying patterns and basic mechanisms of disease, but integrating this new knowledge across scales and applying the results to public health policy are difficult given the non-linear and cross-species interactions inherent to zoonotic infections (13). These complexities can be addressed by harnessing the integrative power and mechanistic insights attainable from analysis of population dynamic models of zoonotic transmission. Here, we review the role of dynamical modeling in the study of zoonoses through an analysis of the current status of the field. Our specific goal is to detect gaps in present knowledge and identify the priorities for future research that will unify, focus, and propel the inter-disciplinary push to combat zoonoses.
A Taxonomy for Zoonotic Dynamics
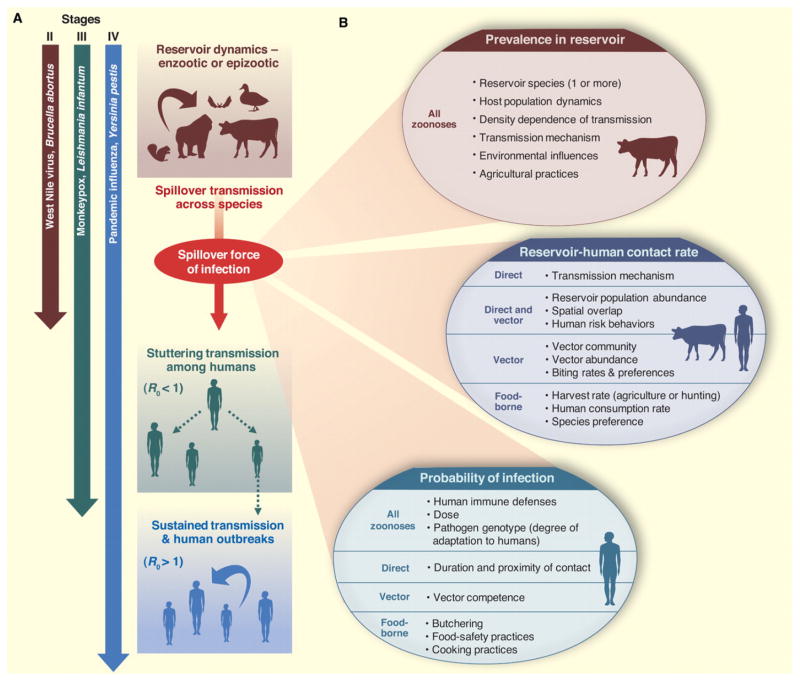
Wolfe et al. (3) proposed a useful classification scheme for pathogens, delineating five stages spanning the range from those exclusively infecting animals (stage I) to those exclusively infecting humans (stage V). The zoonotic component of this scheme (stages II–IV) can be divided into the constituent phases of transmission and associated epidemiological mechanisms (Fig. 1A). Stage II pathogens are those, like West Nile virus or Brucella abortus, that can transmit from animals to humans to cause ‘primary’ infections but do not exhibit human-to-human (‘secondary’) transmission. Stage III pathogens, such as monkeypox virus and Leishmania infantum, spill over into human populations from animal reservoirs and can cause limited cycles of human-to-human transmission that stutter to extinction. Stage IV pathogens persist in animal reservoirs but can cause self-sustaining chains of transmission in human populations; examples include Yersinia pestis (plague) and pandemic influenza. Our approach diverges from Wolfe et al. by basing the distinction among stages II–IV on the basic reproductive number, R0, from the perspective of the human ‘spillover’ hosts. This quantity, defined as the expected number of secondary cases produced by a typical infectious individual in a wholly susceptible population, is a central concept in epidemiological theory (14, 15). R0 enables us to distinguish stages II–IV on dynamical grounds since it quantitatively demarcates pathogens capable of sustained transmission among humans (those with R0>1) from those doomed to stutter to local extinction (R0<1) or those with no onward transmission (R0=0). The dynamics of all zoonoses involve multiple phases, including transmission in the animal reservoir, spillover transmission into humans, and possibly stuttering or sustained transmission among humans. Cross-species spillover transmission is the defining characteristic of a zoonosis, and examination of the factors influencing the force of infection from animals to humans (Fig. 1B) reveals three distinct components: the prevalence of infection in the animal reservoir, the rate at which humans come into contact with these animals, and the probability that humans become infected when contact occurs. These components are each influenced by diverse properties of natural, agricultural and human systems, with important differences driven by the pathogen’s mode of transmission. Significant quantitative or qualitative differences may also arise between zoonoses that use wildlife rather than domesticated animals as reservoirs, owing to differences in frequency, duration and nature of cross-species contacts and in opportunities for human intervention (Fig. 1).
Fig. 1.
(A) Schematic diagram of zoonotic transmission dynamics. Zoonoses can involve as many as four dynamical phases, including enzootic or epizootic circulation in the animal reservoir, spillover transmission from animals to humans, and sometimes self-limiting stuttering chains of human-to-human transmission or sustained transmission leading to outbreaks. Adapting Wolfe et al. (6), we classify zoonotic pathogens into three stages (II, III, and IV) according to their transmissibility among humans. (B) The spillover force of infection is determined by the product of three major components. The force of infection is defined as the per capita rate of infection of susceptible humans. Beneath each major component is a list of contributing factors drawn from many disciplines; these factors may pertain to all zoonoses or to particular transmission modes, as indicated.
Dynamical Models for Zoonoses
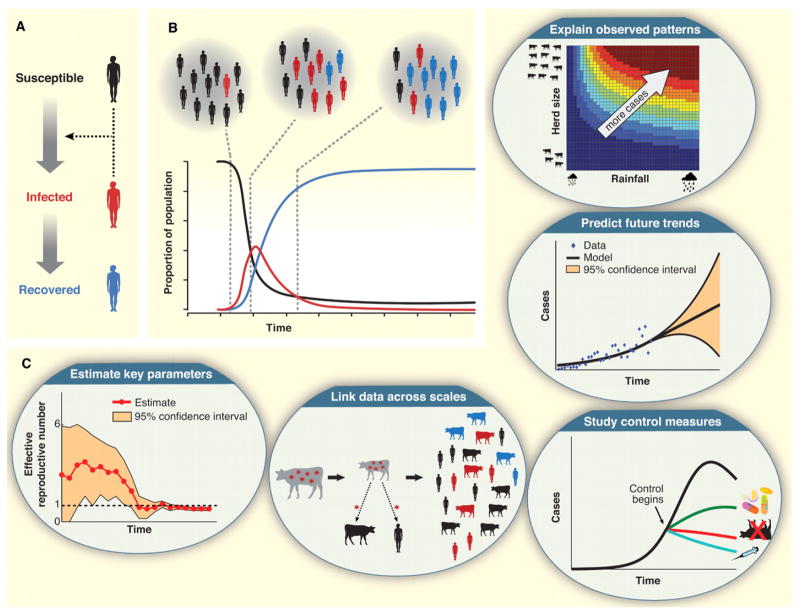
Mathematical models of the population dynamics of infectious diseases (14, 15) use a well-established (and ever-growing) body of theory to construct simplified representations of epidemiological systems. Crucially, dynamical models explicitly represent the key population groups and central processes of epidemic spread. Infectious diseases differ from chronic conditions such as cancer or heart disease, because the risk of infection depends not only on personal risk factors but also upon the state of other individuals in the population. This leads to non-linear interactions among sub-groups in a population that can result in complex and sometimes counter-intuitive epidemic behavior. In the fundamental susceptible-infected-recovered (SIR) model, groups of individuals within the host population are classified as ‘susceptible’ to infection, ‘infected’ and able to transmit the pathogen, or ‘recovered’ and immune to re-infection (Fig. 2A). Transmission of infection to new cases is driven by contacts between susceptible and infectious individuals. Though crude, this model reproduces the classical epidemic curve (Fig. 2B), and has been remarkably successful in elucidating fundamental principles, including the ‘tipping point’ threshold for epidemics to take off if the basic reproduction number, or R0, is greater than 1, and the potential to achieve ‘herd immunity’ through vaccination programs.
Fig. 2.
Dynamical models for epidemics. (A) Schematic diagram of the basic SIR model, showing progression of hosts from susceptible to infected to recovered states. The dotted arrows represent transmission of infection to new cases resulting from contacts between susceptible and infected individuals. (B) The simple epidemic curve (red) predicted by the SIR model for a closed population (i.e., without renewal by immigration or birth of new hosts) and R0 > 1. The curves for the declining proportion of hosts that are susceptible to infection is black and for the rising proportion that are recovered and immune is blue. (C) Dynamical models provide unique insights and allow researchers to ask questions that cannot be addressed by other methods. For example, these models enable estimation of epidemiological parameters linked to key mechanisms, integration of data spanning multiple spatial scales, comparison of alternative control strategies, prediction of future trends, and explanation of observed patterns based on mechanistic hypotheses.
Because of the emphasis on mechanism, dynamical models can address questions outside the scope of statistical and geospatial analyses (Fig. 2C). By adjusting parameter values or reformulating mechanisms, modelers can perform ‘what if’ experiments to study problems that are ethically or logistically unfeasible to study in the real world; for example, the efficacy of different control measures or the population-level consequences of clinical or laboratory findings can be explored. Such cross-scale synthesis also enables researchers to extract key characteristics of epidemics, such as changes in the value of the ‘effective’ reproductive number (Re) as control measures are imposed or the supply of susceptible hosts is depleted. Models can also evaluate the potential influence of unknown information, helping to set priorities for data collection and define the uncertainty associated with model outcomes. Finally, dynamical models can be used to predict future trends of disease spread, although such projections must be accompanied by a comprehensive uncertainty analysis.
Population dynamic modeling has made major contributions to our understanding of zoonotic infections. During the bovine spongiform encephalopathy (BSE) epidemic in Britain, models were used to extract and synthesize basic knowledge from clinical and epidemiological data, while also extrapolating trends from unfolding evidence about the mysterious pathogen (16, 17). Models of rabies transmission have provided biological insight, guided vaccination policy and predicted spatial spread (18–20). Following the emergence of SARS coronavirus (SARS-CoV), models were applied to measure the virus’s transmissibility and refine plans for epidemic containment via infection control and case isolation (21, 22). Modeling studies predicted that quarantine would be a relatively inefficient means of controlling SARS (23, 24), and this was confirmed in later analyses of outbreak data (25).
Comparative analyses have characterized epidemiological phenomena, such as host heterogeneity, across suites of zoonotic pathogens, and have coupled the findings to models to study dynamical consequences (26, 27). For instance, analysis of detailed outbreak data shows that highly infectious ‘superspreaders’ exist for all pathogens (though to varying degrees), and modeling shows that their existence makes outbreaks rarer but more explosive (26). Theoretical studies have illuminated central mechanisms relevant to zoonotic dynamics: analysis of the population dynamics of multi-host pathogens revealed the influence of host species diversity on reservoir dynamics and spillover risk (28), while a hybrid transmission/evolution model showed the potential for stage III zoonoses to adapt to humans before their stuttering chains of transmission die out (29). Zoonotic dynamics have clear parallels with invasion biology (as spillover, stuttering chains and outbreaks correspond roughly to the invasion phases of introduction, establishment, and population expansion) raising the possibility of fruitful cross-fertilization between theoretical frameworks for these fields (30).
In contrast to the complete lack of application of models during the last influenza pandemic in 1968, dynamical models now play a key role in preparing for pandemic influenza strains (e.g., 31). For example, school closure has been identified as an important control measure during the early phase of pandemic spread (32), and was implemented in cities across the USA that were affected by H1N1 influenza (“swine flu”) in spring 2009. Models of both influenza and SARS have exposed the futility of imposing travel restrictions once a pathogen is already spreading within a region (e.g. 24, 33), and such restrictions have been largely avoided following the recognition that the 2009 H1N1 influenza strain was already widespread when it was discovered. Meanwhile, established methods for the estimation of R0 have helped to speed determination of this crucial parameter for the pandemic strain (34).
Surveying the Field: Skews and Gaps
Where has modeling research concentrated across the gamut of zoonotic pathogens and epidemiological challenges? Where are the major gaps in our knowledge, and how can dynamical models be used to integrate empirical findings, guide health policy, and drive innovative research?
We systematically surveyed 442 modeling studies addressing 85 species of zoonotic pathogens, and found surprising gaps and tremendous skews in coverage (for details see Supplementary Online Material). Viral diseases have dominated zoonotic modeling, led by pandemic influenza, SARS and rabies which together account for almost half of all zoonotic models (Table 1; Fig. S1). Bacterial and especially protozoan pathogens have received much less attention relative to their importance. Vector-borne and food-borne zoonoses have been neglected by dynamical modelers compared with infections transmitted by direct contact. Consequently, many zoonotic diseases of great public health concern, such as leptospirosis and yellow fever, have rarely been modeled (Table 1), and so we lack a formal framework to understand the transmission dynamics of these diseases or to respond to sudden changes in their epidemiology. The current literature often fails to account for the multi-host ecology of zoonotic pathogens. The great majority of modeling studies consider just a single phase of the zoonotic process, typically focusing on dynamics in the reservoir or outbreaks in the human population (Fig. S2). Models incorporating spillover transmission—the defining process of zoonotic dynamics—are dismayingly rare. For directly-transmitted zoonoses, we found only six dynamical studies that include a mechanistic model of animal-to-human spillover. For vector-borne and food-borne infections, this number is higher but still a clear minority. Rather than integrating across host species and dynamical phases to address questions aimed at the zoonotic nature of these pathogens, too often we find zoonoses being treated ‘piece-wise’ as a concatenation of single-host processes (or worse, some phases are ignored completely).
Table 1.
Modeling effort for selected zoonotic pathogens, organized by pathogen stage (see Fig. 1A) and number of published dynamical models.
| Pathogen stage | Number of modeling studies | ||||
|---|---|---|---|---|---|
| 0 | 1–5 | 6–10 | 11–20 | >20 | |
| II (spillover only) | Babesia microti Bartonella henselae Chlamydophila psittaci Coxiella burnetii Francisella tularensis Hendra virus Rickettsia prowazekii Rickettsia typhi Streptococcus suis Venezuelan equine EV | Bacillus anthracis Campylobacter jejuni Japanese EV Leptospira interrogans Puumala virus Salmonella typhimurium Tick-borne EV | Brucella abortus Louping ill virus Toxoplasma gondii Trypanosoma brucei rhodesiense West Nile virus | Borrelia burgdorferi Sin Nombre virus Trypanosoma cruzi | BSE Rabies virus |
| III (spillover + stuttering chains) | Andes virus Lassa virus Machupo virus Nipah virus | Leishmania chagasi Crimean-Congo HF virus Monkeypox virus Yersinia enterocolitica | Leishmania infantum | E. coli O157:H7 | Influenza A (avian) Mycobacterium bovis |
| IV (spillover + possible outbreaks) | Barmah forest virus Dengue virus (sylvatic) Leishmania donovani Marburg virus Mayaro virus | Chikungunya virus Ebola virus Ross River virus Yellow fever virus | Yersinia pestis | Influenza A (pandemic) SARS-CoV | |
Abbreviations: EV = encephalitis virus, HF = hemorrhagic fever, BSE = bovine spongiform encephalopathy.
A similar gap is evident in the modeling of stuttering chains of transmission (Fig. S2), wherein zoonotic pathogens transmit inefficiently among humans so any minor outbreaks triggered by spillover events inevitably die out. Despite their limited epidemic potential, such pathogens present epidemiologists with significant challenges that are often best addressed using mathematical models. For instance, monkeypox virus has long been known to spread inefficiently among humans, but its transmissibility appears to be rising as population immunity drops because fewer people have been vaccinated against smallpox (35, 36). Surveillance data for stage III zoonoses, such as monkeypox, Nipah virus or H5N1 avian influenza, can be analyzed to estimate human-to-human transmissibility and to define signatures of possible viral adaptation to humans (37). Given that pathogens in this class are the best-identified threats for future pandemics in the human population, study of their dynamics should be prioritized for attention.
Several patterns stand out among the existing models (Fig. S3). They have been primarily applied to studying the efficacy of control measures, with the secondary aims of estimating epidemiological parameters of interest and explaining observed patterns in field data. Prediction of future trends is a major focus for models of pandemic influenza and BSE, but this aim is rarely applied to other pathogens. Equally notable are the questions missing from these studies. The dynamics of pathogen populations within individual hosts individuals have rarely been included in models of zoonotic transmission dynamics, with the notable exception of food-borne pathogens where the association between pathogen titers in livestock (before and during processing into meat) and infection risk to humans has been studied. Evolutionary issues are similarly neglected, despite the pressing concerns surrounding adaptation to humans and pandemic emergence for several pathogens. It is particularly striking that of the 62 models of SARS dynamics we found, none deals with pathogen evolution, despite the accumulating evidence that the virus was adapting rapidly as it circulated among humans (38). This gap remains because of the paucity of data linking pathogen genotypes to phenotypes at the population scale (in particular transmissibility). This is an important and tractable topic for empirical and theoretical research, especially given the increasing availability of genetic sequence data (39). Interactions among pathogen species have also been largely neglected, even though empirical data show that co-infections are relatively common and that different pathogens can facilitate or hamper each other’s spread through direct or immune-mediated interactions (40). Finally, there has been little research that integrates transmission dynamics of zoonoses with economic considerations, despite the clear relevance of this synthesis to control policy (e.g. 32).
Data and the Link to Reality
A crucial component of a robust and applicable science of zoonotic dynamics is the use of data to estimate parameters and validate model output, and the thoughtful treatment of data-limited situations based on rigorous sensitivity analysis. While most studies use some data to parameterize or validate models (Fig. S3B), their use is highly variable. At the simplest, many authors borrow data-derived parameter values from earlier studies, or fit model projections to epidemic curves. More advanced studies use dynamical reasoning to arrive at new methods of gleaning insights from available data (e.g., 41). For studies aimed at projecting epidemic trends, the gold standard is to validate model output by comparing it with independently gathered data that has not been used for the construction of the model. Notable examples include the ‘post-diction’ of global spread of the 1968 influenza pandemic based on air traffic data (42) and the validation of BSE models developed for England using independent data from Northern Ireland (43). Models can also interact powerfully with population-level experiments to confirm mechanisms underlying observed patterns (44). Broad patterns of data usage are of course determined by the availability of relevant data sets. The free availability of epidemic curves led to two-thirds of SARS modeling studies incorporating population-level data, and early analyses of SARS disease progression enabled widespread use of data-driven parameters (45). For influenza, the well-known clinical course of infection has enabled widespread use of data in parameterizing models, but model-fitting to epidemic curves has been less common. Studies focusing on zoonoses in their animal reservoirs (e.g. rabies and bovine tuberculosis) have relied largely on individual-level parameters, owing to the relative rarity of collated population data for animal diseases. In contrast, however, all the BSE models were fit to population data, reflecting the intensive and cumulative study of the British epidemic and later application of these methods to BSE data from other countries.
Data-free modeling tends to be more common for pathogens that have fewer models overall; this likely reflects either the complete unavailability of data or the lack of opportunity to borrow data-informed parameter values from other studies. However, a broad and vibrant literature has applied statistical methods to analyze the epidemiological and spatial patterns of zoonotic infections (e.g., 46, 47), and opportunities to link these findings with dynamical models too often go unrealized.
Looking beyond the use of data, the best modeling studies are those that engage substantively, and realistically, with current thinking in biology or public health. However, detail and model complexity are not equivalent to realism, and a simple model can yield more insight than a massive simulation that is not fully understood. Detailed simulation models are necessary to address some important questions, but model complexity should be increased cautiously and with an awareness of the associated costs of reduced transparency, a multiplicity of often unknown parameters, and the resulting need for intensive investigation of how model assumptions may influence conclusions (e.g., 31). A hierarchical approach, comparing the behavior of models with different degrees of detail, can aid the design and interpretation of complex models.
Response Dynamics and the Determinants of Effort
Given the prevailing focus of zoonotic models on designing and assessing control measures, and the unique potential of models to estimate important characteristics of epidemics, it is instructive to consider the timeliness with which modelers respond to emerging threats. For zoonotic pathogens that either were newly discovered or have made significant range shifts in recent decades, we constructed temporal profiles of the scientific literature to characterize the dynamics of the modeling community’s response (Figs. 4 and S4). The poster child for rapid response is SARS, for which several modeling analyses were completed within months of the pathogen’s and epidemic’s discovery and were influential in designing outbreak control measures. For Borrelia burgdorferi, the causative agent of Lyme disease, a 10-year delay from discovery to the first model has been followed by intermittent modeling efforts, perhaps reflecting the difficulty of constructing mechanistic models for a vector-borne pathogen with a multi-host sylvatic cycle that is still being characterized, coupled with a vector having a complex life-history incorporating multi-year time delays. West Nile virus exemplifies a different pattern, in which low-level scientific interest suddenly surges following incursions into new geographic territory (wealthy nations in particular), and sporadic modeling efforts follow a few years later.
A sense of urgency and global risk motivates the community to produce and publish dynamical models quickly. Regrettably, for those zoonoses largely restricted to developing countries, such as trypanosomiasis (sleeping sickness), leishmaniasis, and leptospirosis, neglect still applies (Figs. 3B and Table S1) despite their substantial public health impacts. A further, essential factor is the existence of good clinical and epidemiological data to construct and validate models. Here we stress that data must be made publicly available, preferably in real time as occurred for SARS, to maximize both the research opportunity and the resulting public good of scientifically-derived policy. The early availability of genetic sequence data for the current H1N1 influenza pandemic has been exemplary in this regard, but unfortunately the corresponding epidemiological data have been less systematically accessible.
Fig. 3.
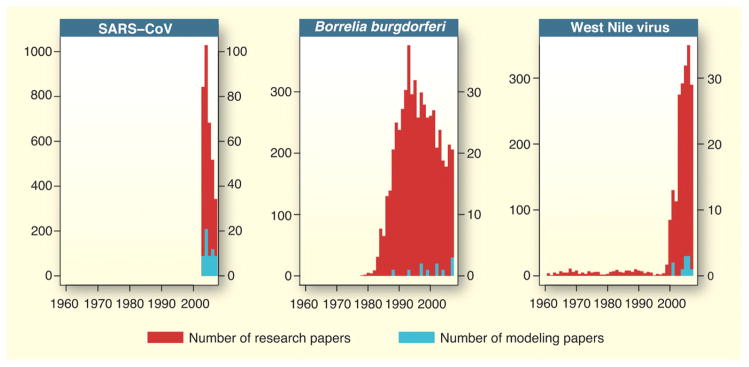
Temporal profiles of total research effort (red) and modeling effort (blue) for recently emerged zoonoses. Figure panels have different y-axis scaling, but in each instance, the scaling for number of modeling studies (right axis) is 1/10th that for the total number of research papers (left axis).
Finally, a key determinant of modeling effort is pathogen life history—in particular the extent to which a pathogen matches the assumptions of the basic SIR family of epidemic models. For infections that are acute, symptomatic and directly-transmitted, models can be constructed (and often parameterized) using ‘off-the-shelf’ techniques, greatly speeding the analysis of newly identified disease threats and enabling extension of the models to address more sophisticated questions. Hence, an important priority for on-going research is to expand the class of pathogens for which there are readily available modeling templates, and worked examples of their connection to datasets, beyond those that conform easily to the standard SIR model.
The Way Forward: Crossing Species, Crossing Disciplines
Significant shortfalls in dynamical studies of particular diseases, as well as entire classes of zoonotic pathogens (notably protozoan and vector-borne infections), can be clearly discerned in the literature (Fig. 3). In contrast, some zoonoses, such as influenza, SARS and BSE, have acted as crucibles for development of new methods for understanding epidemiological complexities, particularly where well-resolved data are available. Unfortunately, even these models have been restricted in scope and there is a need for new models that integrate across phases of zoonotic dynamics and incorporate evolutionary, economic and within-host considerations.
An especially worrying gap is modeling of spillover transmission from animals to humans. The force of infection across species boundaries can be broken into its constituent factors (Fig. 1B), including universal components such as the role of human susceptibility, risk behaviors, and infection prevalence in the reservoir, as well as particular details related to transmission routes and pathogen-specific biology. Spillover risk also can be influenced by dynamical phenomena: one innovative study posits that epizootic peaks of hantavirus in rodent reservoirs, with accompanying spikes in numbers of newly-infected rodents excreting large quantities of virus, lead to high concentrations of virus in the environment and hence an increased risk of human infection (48). Further research will reveal whether such hypotheses may hold true generally across zoonotic systems.
Surprisingly, most of the best models of spillover have been developed for the more complex vector-borne and food-borne zoonoses; this arises because the process of transmission can often be observed directly in such systems. Studies of vector-borne infections have used quantitative frameworks to integrate data on host competency, vector feeding preferences, and environmental conditions to be able to estimate spillover risk from different ecological pathways, though without explicitly incorporating dynamics (49). The food safety literature has treated the risk of pathogens crossing from animal hosts to human exposures in vivid mechanistic detail (50), setting a high standard to be matched by disease ecologists. For directly-transmitted zoonoses, it is straightforward to construct a basic model with cross-species transmission (20), but it is very challenging to delve into underlying mechanisms and estimate key parameters, particularly the cross-species contact rate and the resulting probability of infection, which typically arise from multiple and largely unobserved ecological, behavioral and physiological factors. Nevertheless, the plummeting cost of genetic sequencing brings exciting opportunities for mapping cross-species transmission (51).
A second major gap lies in analysis of stuttering chains of inefficient human-to-human transmission following spillover. Data from these settings are dominated by stochastic effects and heterogeneities among hosts and environments, and analysis is complicated by the fundamental problem of distinguishing between primary and secondary cases. The central challenges in the study of stage III zoonoses include quantifying the rate of human-to-human transmission against a background of spillover, and monitoring for changes in pathogen transmissibility that may represent steps toward emergence of new stage IV pathogens (29).
The study of zoonotic dynamics offers a unique window into fundamental questions of pathogen ecology and evolution, and provides vital insights into public health issues. We need models to reveal the points of vulnerability where intervention against zoonoses will be most effective, and to highlight the gaps in data collection. We need to know when particular zoonotic phases, i.e., reservoir transmission, spillover from animals to humans, stuttering transmission or incipient outbreaks among humans, can be targeted to optimize epidemiological outcomes while reducing cost. How should health policy be adapted to account for environmental change or regional differences in ecology and sociology? How might zoonotic pathogens evolve in response to anthropogenic forces or control strategies? Dynamical models, rooted in data, provide an essential framework for addressing these critical questions.
Supplementary Material
Acknowledgments
This work was supported by the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. We are grateful to Ottar Bjornstad, Ellis McKenzie, Mary Poss, Andrew Read and Lone Simonsen, for valuable comments, and to Benny Gee for assistance with figure preparation. J.L.-S. is grateful for the support of the De Logi Chair in Biological Sciences.
References and Notes
- 1.Woolhouse MEJ, Gowtage-Sequeria S. Emerg Infect Dis. 2005;11:1842. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor LH, Latham SM, Woolhouse MEJ. Phil Trans R Soc Lond B. 2001;356:983. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe ND, Dunavan CP, Diamond J. Nature. 2007;447:279. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens DM, Folkers GK, Fauci AS. Lancet Infect Dis. 2008;8:710. doi: 10.1016/S1473-3099(08)70256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones KE, et al. Nature. 2008;451:990. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer SR, Soulsby EJL, Simpson DIH. Zoonoses: biology, clinical practice, and public health control. Oxford University Press; New York: 1998. [Google Scholar]
- 7.Childs JE. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. 2007;315:389–443. [Google Scholar]
- 8.Weiss RA, McMichael AJ. Nat Med. 2004;10:S70. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smolinksy MS, Hamburg MA, Lederburg J, editors. Microbial threats to health: emergence, detection and response. Institute of Medicine; Washington DC: 2003. [PubMed] [Google Scholar]
- 10.Hassan R, Scholes R, Ash N, editors. Ecosystems and Human Well-being: Current State and Trends. Vol. 1. Island Press; Washington DC: 2005. [Google Scholar]
- 11.Kuiken T, et al. Science. 2006;312:394. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 12.Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Emerg Infect Dis. 2005;11:1822. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daszak P, et al. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. 2007;315:463–475. [Google Scholar]
- 14.Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford University Press; 1991. [Google Scholar]
- 15.Keeling MJ, Rohani P. Modeling infectious diseases in humans and animals. Princeton University Press; Princeton: 2008. [Google Scholar]
- 16.Anderson RM, et al. Nature. 1996;382:779. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson NM, Donnelly CA, Woolhouse MEJ, Anderson RM. Phil Trans R Soc Lond B. 1997;352:803. doi: 10.1098/rstb.1997.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell CA, Smith DL, Childs JE, Real LA. PLoS Biol. 2005;3:382. doi: 10.1371/journal.pbio.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampson K, et al. PLoS Biol. 2009;7:e1000053. doi: 10.1371/journal.pbio.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DL, Lucey B, Waller LA, Childs JE, Real LA. Proc Natl Acad Sci U S A. 2002;99:3668. doi: 10.1073/pnas.042400799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley S, et al. Science. 2003;300:1961. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 22.Lipsitch M, et al. Science. 2003;300:1966. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd-Smith JO, Galvani AP, Getz WM. Proc R Soc Lond B. 2003;270:1979. doi: 10.1098/rspb.2003.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumel AB, et al. Proc R Soc Lond B. 2004;271:2223. [Google Scholar]
- 25.Hsieh YH, et al. J Theor Biol. 2007;244:729. doi: 10.1016/j.jtbi.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Nature. 2005;438:355. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woolhouse MEJ, et al. Proc Natl Acad Sci U S A. 1997;94:338. [Google Scholar]
- 28.Dobson A. Am Nat. 2004;164:S64. doi: 10.1086/424681. [DOI] [PubMed] [Google Scholar]
- 29.Antia R, Regoes RR, Koella JC, Bergstrom CT. Nature. 2003;426:658. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Childs JE, Richt JA, Mackenzie JS. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. 2007;315:1–31. [Google Scholar]
- 31.Halloran ME, et al. Proc Natl Acad Sci U S A. 2008;105:4639. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cauchemez S, et al. Lancet Infect Dis. 2009;9:473. doi: 10.1016/S1473-3099(09)70176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper BS, Pitman RJ, Edmunds WJ, Gay NJ. PLoS Med. 2006;3:845. doi: 10.1371/journal.pmed.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser C, et al. Science. 2009;324:1557. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rimoin AW, et al. Emerg Infect Dis. 2007;13:934. doi: 10.3201/eid1306.061540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fine PEM, Jezek Z, Grab B, Dixon H. Int J Epidemiol. 1988;17:643. doi: 10.1093/ije/17.3.643. [DOI] [PubMed] [Google Scholar]
- 37.Ferguson NM, Fraser C, Donnelly CA, Ghani AC, Anderson RM. Science. 2004;304:968. doi: 10.1126/science.1096898. [DOI] [PubMed] [Google Scholar]
- 38.Sheahan T, et al. J Virol. 2008;82:2274. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grenfell BT, et al. Science. 2004;303:327. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 40.Graham AL, Cattadori IM, Lloyd-Smith JO, Ferrari MJ, Bjornstad ON. Trends Parasitol. 2007;23:284. doi: 10.1016/j.pt.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Wallinga J, Teunis P. Am J Epidemiol. 2004;160:509. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rvachev LA, Longini IM., Jr Math Biosci. 1985;75:3. [Google Scholar]
- 43.Ferguson NM, Ghani AC, Donnelly CA, Denny GO, Anderson RM. Proc R Soc Lond B. 1998;265:545. doi: 10.1098/rspb.1998.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurenson MK, Norman RA, Gilbert L, Reid HW, Hudson PJ. J Anim Ecol. 2003;72:177. [Google Scholar]
- 45.Donnelly CA, et al. Lancet. 2003;361:1761. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King RJ, Campbell-Lendrum DH, Davies CR. Emerg Infect Dis. 2004;10:598. doi: 10.3201/eid1004.030241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostfeld RS, Canham CD, Oggenfuss K, Winchcombe RJ, Keesing F. PLoS Biol. 2006;4:1058. doi: 10.1371/journal.pbio.0040145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauvage F, Langlais M, Pontier D. Epidemiol Infect. 2007;135:46. doi: 10.1017/S0950268806006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Proc R Soc Lond B. 2006;273:2327. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan D, McEwen SA, Lammerding AM, McNab WB, Wilson JB. Prev Vet Med. 1999;41:55. doi: 10.1016/s0167-5877(99)00032-x. [DOI] [PubMed] [Google Scholar]
- 51.Song HD, et al. Proc Natl Acad Sci U S A. 2005;102:2430. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.