Highlights
- •
SARS-CoV recognizes host receptor ACE2 via its receptor-binding domain (RBD).
- •
Residue differences in host ACE2 present species barriers for SARS-CoV infections.
- •
SARS-CoV can adapt to ACE2 from several species through RBD mutations.
- •
Structural studies have revealed receptor adaptation mechanisms of SARS-CoV.
- •
Structural studies also allow predictions of future SARS-CoV evolution.
Keywords: Coronavirus, Spike protein, Severe acute respiratory syndrome, Middle East respiratory syndrome, Virus evolution
Abstract
Receptor recognition is a major determinant of the host range, cross-species infections, and pathogenesis of the severe acute respiratory syndrome coronavirus (SARS-CoV). A defined receptor-binding domain (RBD) in the SARS-CoV spike protein specifically recognizes its host receptor, angiotensin-converting enzyme 2 (ACE2). This article reviews the latest knowledge about how RBDs from different SARS-CoV strains interact with ACE2 from several animal species. Detailed research on these RBD/ACE2 interactions has established important principles on host receptor adaptations, cross-species infections, and future evolution of SARS-CoV. These principles may apply to other emerging animal viruses, including the recently emerged Middle East respiratory syndrome coronavirus (MERS-CoV). This paper forms part of a series of invited articles in Antiviral Research on “From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses”.
1. SARS coronavirus
The severe acute respiratory syndrome (SARS) epidemic broke out in 2002–2003 in southern China, and spread to other regions of Asia and also to Europe and North America (Lee et al., 2003, Yu et al., 2004). It caused more than 8000 infections worldwide with an approximately 10% fatality rate, along with huge economic losses. SARS patients developed flu-like symptoms, often followed by acute atypical pneumonia and respiratory failure. The epidemic was eventually controlled by conventional public health measures. SARS briefly re-emerged in 2003–2004, causing 4 sporadic infections but no fatality or human-to-human transmission (Liang et al., 2004, Song et al., 2005). Since then, there have been no reported cases of naturally occurring SARS. Because of the dramatic fashion how it emerged and disappeared, SARS presents a unique and puzzling case in the history of human epidemics.
A novel coronavirus, SARS coronavirus (SARS-CoV), was the etiological agent of SARS (Ksiazek et al., 2003, Peiris et al., 2003). Coronaviruses are a family of common, ancient, and diverse viruses that infect many mammalian and avian species (Perlman and Netland, 2009). Coronavirus virions contain an envelope, a helical capsid, and a single-stranded and positive-sense RNA genome with a length of 27–32 kb; their genomes are the largest among all RNA viruses. They were named “coronavirus” because of the large spike protein molecules on the virus surface that give the virions a crown-like shape (corona in Latin means crown) (Fig. 1 A, B). Coronaviruses can be classified into at least three major genera, α, β, and γ (Gonzaalez et al., 2003). Six coronaviruses are currently known to infect humans. The first five are NL63 and 229E from α-genus, and OC43, HKU1, and SARS-CoV from β-genus (Perlman and Netland, 2009). A new member of β-genus, the Middle East respiratory syndrome coronavirus (MERS-CoV), has recently emerged from the Middle East and infected at least 88 people with a mortality rate of more than 50% (de Groot et al., 2013, Holmes and Dominguez, 2013, Zaki et al., 2012). In the past coronaviruses were considered to be mild human viral pathogens that only cause self-resolving respiratory diseases in humans. The emergence of SARS-CoV and MERS-CoV has changed this conception.
Fig. 1.
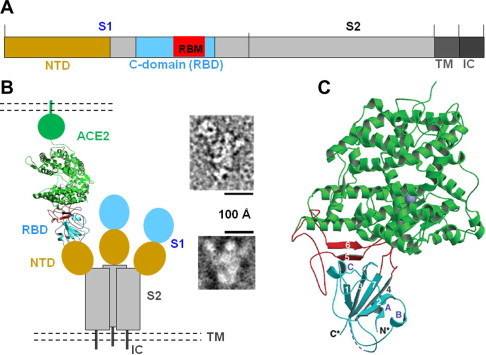
Structure of the SARS-CoV spike protein complexed with its receptor ACE2. (A) Schematic domain structure of SARS-CoV spike protein. NTD: N-terminal domain. RBM: receptor-binding motif. TM: transmembrane anchor. IC: intracellular tail. (B) Overall structure of trimeric SARS-CoV spike protein complexed with ACE2, including both schematic topology of the spike protein (left) and negative-stain electron micropic images of the spike protein ectodomain with (upper right) or without a bound ACE2 (lower right). (C) Crystal structure of SARS-CoV RBD (i.e., S1 C-domain) complexed with ACE2. ACE2 is in green, RBD core structure in cyan, and RBM in red. Figure adapted from (Li et al., 2006a, Li et al., 2005a).
SARS-CoV infects a wide variety of animal species, including humans, palm civets, monkeys, domestic cats, ferrets, hamsters, raccoon dogs, and bats (Li et al., 2006c) (Fig. 2 A). It is believed that bats harbor the precursors of SARS-CoV and thus are the natural reservoir for the virus (Lau et al., 2005, Li et al., 2005b). In addition, palm civets played a key role in the spread of SARS-CoV to humans and thus were the interim reservoir or amplifying host for the virus (Gonzaalez et al., 2003, Li et al., 2005b, Li et al., 2006c, Shi and Hu, 2008). Not all animals can be efficiently infected by SARS-CoV. For example, rats are resistant to SARS-CoV infections, while mice can be infected by SARS-CoV only inefficiently (Li et al., 2004, Subbarao et al., 2004). On the other hand, various strains of SARS-CoV have been isolated from different hosts at different times (Fig. 2B). These SARS-CoV strains include hTor02 (human strain isolated during the 2002–2003 SARS outbreak) (Marra et al., 2003), cSz02 (civet strain isolated from marketplace palm civets during the 2002–2003 SARS outbreak) (Guan et al., 2003), hcGd03 (human and civet strain isolated during the 2003–2004 sporadic SARS infections) (Song et al., 2005), and cGd05 (civet strain isolated from wild civets in 2005) (Liu et al., 2007), hHae08 (human strain after adaptation to cultured human cells) (Sheahan et al., 2008b), and bHKU05 (bat strain isolated in 2005) (Lau et al., 2005). The large number of hosts and identification of the viral strains from different hosts make SARS-CoV an excellent model system for understanding host ranges and cross-species infections of emerging animal viruses.
Fig. 2.
Receptor and RBD residues that play important roles in the host range and cross-species infections of SARS-CoV. (A) Alignment of ACE2 residues from different animal species that are critical for the host range of SARS-CoV. The GenBank accession numbers are AY623811 (human ACE2), AY881174 (civet ACE2), AY881244 (rat ACE2), EF408740 (mouse ACE2), and GQ999937 (bat ACE2). ∗ refers to an N-linked glycosylation site at the indicated position. (B) Alignment of RBD residues from different SARS-CoV strains that have undergone naturally selected mutations. The GenBank accession numbers are AAP41037 (hTor02 spike), AY304488 (cSz02 genome), AAS10463 (hcGd03 spike), ABF68956 (cHb05 spike), and AAY88866 (bHKU05 spike). (C) Distribution of the above RBD residues at the interface of SARS-CoV RBD and ACE2. Figure adapted from (Li, 2008, Wu et al., 2012).
2. Spike protein and receptor
The envelope-anchored trimeric spike protein mediates coronavirus entry into host cells by binding to its host receptor and subsequently fusing host and viral membranes (Bosch et al., 2003, Gallagher and Buchmeier, 2001). The spike protein consists of three segments, an ectodomain, a single-pass membrane anchor, and a short intracellular tail (Fig. 1A). The ectodomain can be divided into a receptor-binding S1 subunit and a membrane-fusion S2 subunit. S1 contains two independent domains, an N-terminal domain (NTD) and a C-domain (Li, 2012). Depending on the virus, either NTD or C-domain (occasionally both) binds to a host receptor and functions as a receptor-binding domain (RBD) (Breslin et al., 2003, Godet et al., 1994, Krempl et al., 1997, Kubo et al., 1994, Lin et al., 2008). Coronaviruses recognize a variety of cell-surface molecules as their host receptors, including proteins, sugars, and heparan sulfate (Perlman and Netland, 2009). How they evolved to do so has been revealed by structural studies of coronavirus RBD/receptor interactions (Chen and Li, 2013, Chen et al., 2012, Li, 2012, Li et al., 2005aa; Peng et al., 2011, Peng et al., 2012, Reguera et al., 2012, Wu et al., 2011, Wu et al., 2009). The S1 C-domain of SARS-CoV is the RBD that recognizes host angiontensin-converting enzyme 2 (ACE2) as its receptor (Babcock et al., 2004, Li et al., 2003, Sui et al., 2004, Wong et al., 2004, Xiao et al., 2003). The SARS-CoV RBD is also a major target of neutralizing antibodies (He et al., 2006, He et al., 2005, He et al., 2004, Huang et al., 2006, Prabakaran et al., 2006). ACE2 is a zinc-dependent peptidase that functions in the renin-angiotensin pathway and regulates blood pressure (Donoghue et al., 2000, Yagil and Yagil, 2003). However, the physiological function of ACE2 is not related to its role as the SARS-CoV receptor (Li et al., 2005b). ACE2 contains an N-terminal peptidase domain and a C-terminal collectrin domain. The peptidase domain has a claw-like structure with two lobes. The enzymatic active site of ACE2 is buried in a deep cavity between the two lobes (Towler et al., 2004). The binding interactions between SARS-CoV RBD and ACE2 largely determine the host range and cross-species infections of SARS-CoV and hence are the focus of this review.
Several lines of research have established receptor recognition as one of the major species barriers between humans and animals for SARS-CoV infection. That is, whether a host is susceptible to SARS-CoV is largely determined by the binding affinity between SARS-CoV RBD and host ACE2 in the initial step of viral attachment. First, epidemiologic and biochemical studies show that the infectivity of different SARS-CoV strains in host cells is positively correlated with the binding affinity between the RBD of each strain and ACE2 expressed by the host cell (Li et al., 2005b, Qu et al., 2005, Song et al., 2005). Second, cell biology studies show that SARS-CoV infects mouse cells inefficiently because of the poor binding affinity between murine ACE2 and SARS-CoV. In contrast, mouse cells that express human ACE2 on their surface are highly susceptible to SARS-CoV (Moore et al., 2004). Moreover, currently known bat SARS-CoV strains do not infect human cells (Ren et al., 2008). However, when their RBD is replaced with the RBD from a human SARS-CoV strain, the synthetic recombinant bat SARS-CoV strain infects human cells efficiently (Becker et al., 2008). Third, animal studies show that mice are not effective hosts for SARS-CoV, but transgenic mice that express human ACE2 become good hosts for SARS-CoV (McCray et al., 2007, Tseng et al., 2007). Fourth, reverse genetics studies show that synthetically reconstructed civet SARS-CoV strains depend completely on ACE2 for entry and infection (Sheahan et al., 2008a, Sheahan et al., 2008b). Therefore, all evidence so far has identified receptor recognition as an important limiting step for the cross-species infections of SARS-CoV.
3. Structure of spike protein/receptor complex
In a series of studies, my colleagues and I have investigated the interactions between the SARS-CoV spike protein and ACE2 via a combination of electron microscopic and X-ray crystallographic approaches. We first collected negative-stain electron microscopic images of the SARS-CoV spike protein ectodomain, and showed that it is a clove-shaped trimer with three individual S1 heads and a trimeric S2 stalk (Fig. 1B) Li et al., 2006a). ACE2 binds to the tip of the trimeric spike protein ectodomain where the S1 C-domain is located. S1 NTD is probably underneath the C-domain. These data motivated us to construct a schematic model of the SARS-CoV invasion machinery to demonstrate the spatial relationships among the different parts of the spike protein and receptor (Fig. 1B). We then determined the crystal structure of SARS-CoV RBD complexed with human ACE2 (Fig. 1C) (Li et al., 2005a, Li et al., 2006b). The structure shows that SARS-CoV RBD contains two subdomains, a core structure and an extended loop. The core structure is a five-stranded anti-parallel β-sheet, with several short connecting α-helices. The extended loop lies on one edge of the core structure, and presents a gently concave surface to interact with the N-terminal lobe of the ACE2 peptidase domain. Because this extended loop makes all the contact with ACE2, it has been termed “receptor-binding motif” (RBM) (Fig. 2C) (Li et al., 2005a). Overall, 14 RBM residues directly contact 18 ACE2 residues. These structural studies have provided a structural platform for understanding how the spike/ACE2 interactions determine the cross-species infections and human-to-human transmission of SARS-CoV.
4. Civet-to-human jump and human-to-human transmission
To understand the civet-to-human jump and human-to-human transmission of SARS-CoV, we determined more crystal structures of the interfaces between various SARS-CoV strains and human (hACE2) or civet ACE2 (cACE2) (Fig. 3 ) (Li, 2008). Residue 479 is an asparagine in human epidemic viral strain hTor02, but a lysine in civet viral strain cSz02 (Figs. 2B, 3A, C). hACE2 Lys31 forms a salt bridge with hACE2 Glu35 in a hydrophobic environment (Fig. 3A). cSz02 Lys479 would competes with Lys31 for Glu35 as a salt bridge partner, destabilizing the binding interface. Hence viral mutation K479N increases the RBD/hACE2 binding affinity. The reason why cACE2, not hACE2, can accommodate Lys479 is because cACE2 Thr31 cannot form a salt bridge with Glu35, making Glu35 available to form a salt bridge with cSz02 Lys479 (Fig. 3C). Thus civet viral strains with Lys479 have high affinity for cACE2, but not hACE2. Therefore, mutation K479N in SARS-CoV RBD played an important role in the civet-to-human jump of SARS-CoV.
Fig. 3.
Structural basis for the civet-to-human jump and human-to-human transmission of SARS-CoV. (A)–(D) Structural details of the interfaces between ACE2 from human or civet and RBD from different viral strains. Figure adapted from (Li, 2008).
On the other hand, residue 487 is a threonine in hTro02, but a serine in cSz02 (Figs. 2B, 3B, D). Lys353 forms a salt bridge with hACE2 Asp38 in a hydrophobic environment. Formation of the salt bridge requires support from the methyl group of Thr487 (Fig. 3B). Hence mutation T487S decreases the RBD/hACE2 affinity. The low-pathogenicity human viral strain hcGd03 contains Ser487, explaining why it had failed to transmit from human to human. In comparison, cACE2 Glu38 has a longer side chain than hACE2 Asp38, and can form a salt bridge with Lys353 in the absence of the support from residue 487 (Fig. 3D). Thus civet viral strains with Ser487 can transmit from civets to civets, but not from humans to humans. Therefore, mutation S487T in SARS-CoV RBD facilitated human-to-human transmission of SARS-CoV.
Overall, our studies suggest that two residue changes between civet and human ACE2, T31K and E38D, present major species barriers between the two species for SARS-CoV infections (Fig. 3A). These critical residue changes occur at or near two virus-binding hotspots on ACE2, one centering on residue 31 (hotspot-31) and the other on residue 353 (hotspot-353). As a result, early civet SARS-CoV isolates were unable to infect human cells because they were not adapted to the two virus-binding hotspots on human ACE2, and SARS-CoV has evolved to gain sustained infectivity for human cells through stepwise adaptation mutations at residues 479 and 487 in its RBD. Mutation at residue 479 likely determines whether SARS-CoV can infect humans, whereas mutation at residue 487 largely determines whether SARS-CoV can maintain sustained infections in humans.
5. Adaptations to human or civet receptor
The structural analysis above has focused on two RBM mutations, K479N and S487T, which played critical roles in SARS-CoV’s evolution to overcome major species barriers between palm civets and humans for cross-species infections. However, a number of other RBM mutations have been isolated from different SARS-CoV strains, and these mutations are distributed at 5 positions in the RBM: 442, 472, and 480, as well as 479 and 487 (Fig. 2B, C). To examine why these RBM mutations were naturally selected, we investigated the impact of each mutation on receptor binding using a combination of biochemical, virological, and crystallographic methods (Wu et al., 2012). Using surface plasmon resonance and pseudotyped viral infection assays, we found that each of these mutations enhances the binding affinity of RBD to either human ACE2 or civet ACE2 (Fig. 4 A). By determining the crystal structures of mutant SARS-CoV RBDs complexed with human or civet ACE2, we showed that these mutations all surround the two virus-binding hotspots on ACE2, hotspot-31 and hotspot-353 (Fig. 4B). As discussed earlier, both of these hotspots consist of a critical salt bridge that is buried in a hydrophobic environment (Fig. 4B). In this environment, the salt bridges not only provide a significant amount of energy for the virus/receptor binding interaction, but also fill critical voids in the hydrophobic stacking interactions at the virus/receptor binding interface. Consequently, each of the RBM mutations in different SARS-CoV strains enhances viral interactions with either human or civet ACE2 by strengthening the hotspot structures (Fig. 4A). For example, residue 31 is a lysine in human ACE2, and RBD residue Tyr442 has partial steric clash with the side chain of Lys31 and thereby decreases viral binding affinity with human ACE2 (Figs. 3A, 4B). RBD residue Phe442 relieves this steric clash and strengthens the structure of hotspot-31, and thus is a viral adaptation to human ACE2. On the other hand, residue 31 is a threonine in civet ACE2, and RBD residue Tyr442 forms a hydrogen bond with Thr31 and thereby increases viral binding affinity with civet ACE2 (Figs. 3C, 4B). Thus RBD residue Tyr442 is a viral adaptation to civet ACE2. Similarly, we have elucidated the structural basis for all of the other RBM mutations that were viral adaptations to either human or civet ACE2 (Fig. 4A). These studies elucidate comprehensive and detailed mechanisms for SARS-CoV adaptations to human or civet receptor, and provide valuable knowledge about the evolutionary strategies that viruses may take for host receptor adaptations.
Fig. 4.
Structural basis for SARS-CoV adaptations to human or civet receptor. (A) Summary of host receptor adaptation by SARS-CoV. Listed are adaptations of RBM residues to human or civet ACE2. Arrows point from less well adapted residues to better adapted residues. Double arrows connect equally well adapted residues. (B) Detailed structure of the hTor02 RBD/human ACE2 interface. ACE2 residues are in green, SARS-CoV residues that underwent mutation are in magenta, and SARS-CoV residues that played significant roles in the mutations are in cyan. Figure adapted from (Wu et al., 2012).
6. Major species barriers between humans and mice, rats or bats
We further extended our structural analysis to the interactions between SARS-CoV RBD and ACE2 from mouse, rat and bat. Using human ACE2 as the reference molecule, we identified key residue changes in these ACE2 molecules that disfavor SARS-CoV binding (Fig. 2A, Fig. 5 ). First, mouse ACE2 contains a critical K353H residue change. A histidine at the 353 position of ACE2 does not fit into the virus/receptor binding interface as well as a lysine (Fig. 5A). Consequently, mice support SARS-CoV infections inefficiently. Second, rat ACE2 contains two residue changes that disfavor SARS-CoV binding. In addition to the K353H residue change as in mouse ACE2, rat ACE2 also contains an M82N residue change, which introduces a glycosylation site. A glycan at the 82 position of ACE2 would lead to steric interference with SARS-CoV binding (Fig. 5B). Consequently, rats are resistant to SARS-CoV infections. Third, a number of ACE2 molecules have been isolated from different bat species with diverse amino acid sequences (Hou et al., 2010). None of these bat ACE2 molecules functions as an effective receptor for currently known SARS-CoV strains. Among them, ACE2 from Daubenton’s bat supports SARS-CoV infections at low levels. Compared with human ACE2, Daubenton’s bat ACE2 contains three critical residue changes, K31N, E35K, and Y41H. The K31N and E35K residue changes may turn the K31-E35 salt bridge to a weaker N31-K35 hydrogen bond (Fig. 5C), whereas the Y41H residue change likely weakens the support for the K353-D38 salt bridge because histidine is a poorer hydrophobic stacker than tyrosine (Fig. 5D). Thus these residue changes in Daubenton’s bat ACE2 decrease the RBD/ACE2 binding affinity. Overall, our studies have identified critical residue changes between human ACE2 and mouse, rat, or bat ACE2 that serve as major species barriers for SARS-CoV infections.
Fig. 5.
Structural basis for the major species barriers between humans and mice, rats, or bats for SARS-CoV infections. (A)–(D) Structural details of the interfaces between ACE2 from mouse, rat or bat and RBD from human SARS-CoV strain hTor02. Figure adapted from (Hou et al., 2010, Li et al., 2005a).
7. Adaptations to mouse receptor
SARS-CoV adaptations to mouse ACE2 have been explored (Frieman et al., 2012, Roberts et al., 2007). Although human SARS-CoV strains did not infect mice efficiently, they could adapt to mice through serial passage in the respiratory tract of young mice and cause lethal diseases. A number of mutations were identified in these mouse-adapted SARS-CoV strains. Nearly all of these mutations are located in the RBM region of the spike protein and surround the two virus-binding hotspots on mouse ACE2. Although biochemical and structural studies have not been done to characterize the impact of these mutations on receptor binding, these mutations presumably enhance the binding of the RBD to mouse ACE2. For example, a Y436H mutation repeatedly emerged during SARS-CoV adaptations to mice. At the interface of hTor02 RBD and human ACE2, RBD Tyr436 forms a hydrogen bond with hACE2 Asp38, providing support to the K353-D38 salt bridge at hotspot-353 (Fig. 4B). In mouse ACE2, residue 353 is a histidine, which is unlikely to form a salt bridge with Asp38. Instead, RBD His436 in the mouse-adapted SARS-CoV strains may form a strong hydrogen bond with Asp38, partially compensating for the loss of the K353-D38 salt bridge. Detailed mechanisms of these mouse-adapted SARS-CoV mutations still wait to be characterized by structural and biochemical studies.
8. Host range and cross-species infections
The structural analysis reviewed above allowed us to summarize the host range and cross-species infections of SARS-CoV (Fig. 6 ). Bat cells do not support efficient infections of civet or human SARS-CoV strains because of residue changes (e.g. K31N, E35K, and Y41H) in their ACE2. However, given the diversities of bat ACE2 molecules, it is possible that future studies will identify a bat ACE2 that supports SARS-CoV infections. On the other hand, currently known bat SARS-CoV strains cannot jump from bats to civets or humans because of truncations in their RBMs (Fig. 2B). Significant evolution would be needed for these RBMs to acquire sufficient binding affinity for civet or human ACE2. Once SARS-CoV had jumped from bats to civets, it underwent further mutations in civets. The first K479N mutation allowed SARS-CoV to jump from civets to humans and the second S487T mutation allowed SARS-CoV to transmit from human to human, leading to the severe SARS outbreak in 2002–2003. In the following year, SARS-CoV only acquired the K479N mutation, but not the S487T mutation, leading to the sporadic SARS infections with no human-to-human transmission. Mice support SARS-CoV infection at low levels because of the K353H residue change in their ACE2, and rats are resistant to SARS-CoV infection because of the K353H and M82N double residue changes. In sum, critical residue changes in animal ACE2 molecules present species barriers for SARS-CoV infections. Through natural evolutions of its RBD at key positions, SARS-CoV overcame the species barriers between some animal species (e.g., between bats and civets and between civets and humans), but not others (e.g., between civets and mice or between civets and rats).
Fig. 6.
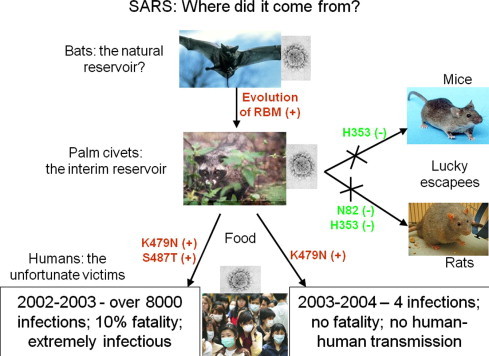
Summary of host range and cross-species infections of SARS-CoV. RBD mutations that overcame species barriers for the cross-species infections of SARS-CoV are in red and labeled as “+”. Residue changes in ACE2 that form species barriers and disfavor the cross-species infections of SARS-CoV are in green and labeled as “−”.
9. Structure-based prediction of future evolution
Predictions of future evolution of a virus are a difficult, if not completely impossible, task. However, our detailed structural analysis of the host receptor adaptation mutations in SARS-CoV RBD has allowed us to predict, design, and test optimized SARS-CoV RBDs that may resemble future evolved forms of the virus (Fig. 7 ) (Wu et al., 2012). For example, a form of RBD optimized to bind human ACE2 (human-optimized form) contains all of the hACE2-adapted residues (Phe-442, Phe-472, Asn-479, Asp-480, and Thr-487). It has exceptionally high affinity for hACE2, but lower affinity for cACE2. A form of RBD optimized to bind civet ACE2 (civet-optimized form) contains all of the cACE2-adapted residues (Tyr-442, Pro-472, Arg-479, Gly-480, and Thr-487). It possesses exceptionally high affinity for cACE2, and importantly, also harbors substantial affinity for hACE2. To our knowledge, this was the first and has been the only case of knowledge-based prediction of viral evolution that leads to improved receptor-binding affinities.
Fig. 7.
Structure-based prediction of future SARS-CoV evolution. Listed are SARS-CoV strains, critical RBD residues, and binding affinities for human or civet ACE2. Arrows suggest directions of SARS-CoV evolution.
Will SARS come back? If it does, in what form will it be back? Our studies suggest that if SARS-CoV were given the opportunity to keep infecting humans and evolving in human cells, hTor02 RBD might evolve into the human-optimized form by acquiring two mutations at the 442 and 472 positions (Fig. 7). Luckily, however, SARS-CoV has been absent in humans since 2003–2004. Nevertheless, SARS-CoV could still be infecting and evolving in wild palm civets. In fact, the cHb05 strain was isolated from wild palm civets in 2005 (Liu et al., 2007), two years after the SARS epidemic. Additionally, the cHb05 strain appears to be significantly better adapted to civet ACE2 than the cSz02 strain isolated during the SARS epidemic, having acquired mutations at positions 472, 479, and 480 (Fig. 2B, Fig. 4A, Fig. 7). Alarmingly, the cHb05 strain is only one mutation (S487T) away from evolving into the civet-optimized form with substantial affinity for human ACE2 (Fig. 7). Overall, our research provides a molecular and structural blueprint for tracking and predicting future SARS-CoV evolution in animals, which helps prevent and control potential future SARS outbreaks in the human population.
10. Comparison with MERS coronavirus
The newly emerged MERS-CoV shares clinical and genetic features with SARS-CoV. Like SARS-CoV, MERS-CoV often causes acute pneumonia and renal failure in patients. In addition, MERS-CoV also belongs to the β-genus and its S1 C-domain has been identified to be the RBD (Du et al., 2013, Mou et al., 2013). But unlike SARS-CoV, MERS-CoV uses dipeptidyl peptidase 4 (DPP4) as its receptor (Raj et al., 2013). Crystal structures of MERS-CoV RBD have recently been determined, either by itself or in complex with its receptor DPP4 (Chen et al., 2013, Lu et al., 2013, Wang et al., 2013). MERS-CoV RBD also contains a core structure and an accessory subdomain, the latter of which functions as RBM. The core structures of MERS-CoV and SARS-CoV RBDs are highly similar, but their RBMs are markedly different, resulting in recognition of different receptors. Like SARS-CoV, MERS-CoV also likely originated from bats because the genomic sequence of MERS-CoV is closely related to those of certain bat coronaviruses (Annan et al., 2013, Holmes and Dominguez, 2013, Lau et al., 2013). Moreover, MERS-CoV is able to infect both human and bat cells and appears to recognize both human and bat DPP4 molecules. These findings suggest that MERS-CoV may be able to transmit from bats to humans and then from human to human with little or no adaptation in its RBD. Details of the host range and cross-species infections of MERS-CoV will need to be characterized by future genetic, biochemical, and structural studies.
11. Concluding remarks
Research on SARS-CoV/receptor interactions has elucidated the structural basis for the cross-species infections and human-to-human transmission of SARS-CoV, and has also provided a structural framework for monitoring and predicting future SARS-CoV evolution in animals. These studies have established several important principles on host receptor adaptations and cross-species infections of viruses. First, one or a few seemingly trivial mutations at the receptor-binding surface of a virus may lead to dramatic epidemic outcomes by facilitating cross-species infections and human-to-human transmission of the virus. Second, one or a few residue changes in the virus-binding region of a host receptor protein may determine the host’s fate and role in a viral epidemic by presenting species barriers for viral infections. Third, predictions of future viral evolution may be possible if virus/receptor interactions can be understood in great structural detail. These principles may apply to other emerging animal viruses, including the newly emerged MERS-CoV.
What are the remaining important questions regarding the host range and cross-species infections of SARS-CoV? First, what receptor do currently known bat SARS-CoV strains use when they infect bat cells? To solve this problem, it will be important to determine whether the bat receptor is an unidentified form of bat ACE2 or an entirely new receptor. Second, what is the detailed structural mechanism by which bat SARS-CoV strains adapted to civet ACE2 for the critical bat-to-civet jump? It is possible that none of the currently known bat SARS-CoV strains was able to adapt to civet or human ACE2 owing to the significant differences between their RBDs and the RBDs from the civet or human SARS-CoV strains. Thus, currently known bat SARS-CoV strains were unlikely to be the immediate precursors of the civet or human SARS-CoV strains that caused the 2002–2003 SARS epidemic. Instead, there may exist one or several hitherto unidentified bat SARS-CoV strains that not only use bat ACE2 as their receptor, but can also adapt to and recognize civet ACE2 with relative ease. Hence, finding these bat SARS-CoV strains will fill in an important missing link associated with the SARS epidemic. In sum, as the natural reservoir for SARS-CoV, MERS-CoV and many other coronaviruses, bats should be carefully studied for how they support coronavirus infections and how coronaviruses may jump from bats to other animal species and pose threats to humans.
Acknowledgments
This work was supported in part by NIH grant R01AI089728. Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute.
References
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Esshaki D.J., Thomas W.D., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Nat. Acad. Sci. U.S.A. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin J.J., Mork I., Smith M.K., Vogel L.K., Hemmila E.M., Bonavia A., Talbot P.J., Sjostrom H., Noren O., Holmes K.V. Human coronavirus 229E: Receptor binding domain and neutralization by soluble receptor at 37 degrees C. J. Virol. 2003;77:4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li F. Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J. Virol. 2013;87:4118–4120. doi: 10.1128/JVI.03476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lin Y.L., Peng G.Q., Li F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Nat. Acad. Sci. U.S.A. 2012;109:17966–17971. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.L., Baric R.S., Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R.J., Baker, S.C., Baric, R.S., Brown, C.S., Drosten, C., Enjuanes, L., Fouchier, R.A., Galiano, M., Gorbalenya, A.E., Memish, Z., Perlman, S., Poon, L.L., Snijder, E.J., Stephens, G.M., Woo, P.C., Zaki, A.M., Zambon, M., Ziebuhr, J., 2013. Middle East Respiratory Syndrome Coronavirus (MERS-CoV); Announcement of the Coronavirus Study Group. Journal of Virology Published online ahead of print. [DOI] [PMC free article] [PubMed]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K., Lu L., Wang L., Debnath A.K., Zheng B.J., Zhou Y., Jiang S. Identification of Receptor-Binding Domain in S protein of the Novel Human Coronavirus MERS-CoV as an Essential Target for Vaccine Development. J. Virol. 2013;87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (Coronavirus) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- He Y.X., Li J.J., Li W.H., Lustigman S., Farzan M., Jiang S.B. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006;176:6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- He Y.X., Lu H., Siddiqui P., Zhou Y.S., Jiang S.B. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- He Y.X., Zhou Y.S., Wu H., Luo B.J., Chen J.M., Li W.B., Jiang S.B. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: Implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Holmes K.V., Dominguez S.R. The New Age of Virus Discovery: Genomic Analysis of a Novel Human Betacoronavirus Isolated from a Fatal Case of Pneumonia. MBio. 8. 2013;4:e00548-12. doi: 10.1128/mBio.00548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.X., Peng C., Yu M., Li Y., Han Z.G., Li F., Wang L.F., Shi Z.L. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W.H., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S.X., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of Neutralizing Epitopes and the Receptor-Binding Site within the Amino-Terminal 330 Amino-Acids of the Murine Coronavirus Spike Protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus hku5 in japanese pipistrelle: implications for the origin of the novel middle east respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Nat. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Berardi M., Li W.H., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W.H., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li F., Li W.H., Farzan M., Harrison S.C. Interactions between SARS coronavirus and its receptor, Nidoviruses: toward Control of Sars and Other Nidovirus Diseases. Adv. Exp. Med. Biol. 2006;581:229–234. doi: 10.1007/978-0-387-33012-9_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.D., Shi Z.L., Yu M., Ren W.Z., Smith C., Epstein J.H., Wang H.Z., Crameri G., Hu Z.H., Zhang H.J., Zhang J.H., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S.Y., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W.H., Greenough T.C., Moore M.J., Vasilieva N., Somasundaran M., Sullivan J.L., Farzan M., Choe H. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 2004;78:11429–11433. doi: 10.1128/JVI.78.20.11429-11433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Moore M.J., Vasilieva N., Sui J.H., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G.D., Chen Q.X., Xu J.G., Liu Y.F., Lim W., Peiris J.S.M., Anderson L.J., Ruan L., Li H., Kan B., Di B., Cheng P., Chan K.H., Erdman D.D., Gu S.Y., Yan X.G., Liang W.L., Zhou D.H., Haynes L., Duan S.M., Zhang X., Zheng H., Gao Y., Tong S.X., Li D.X., Fang L., Qin P.Z., Xu W.B. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg. Infect. Dis. 2004;10:1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.X., Fen Y., Wong G., Wang L.P., Li B., Zhao X.S., Li Y., Smaill F., Zhang C.S. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J. Gen. Virol. 2008;89:1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- Liu L., Fang Q., Deng F., Wang H.Z., Yi C.E., Ba L., Yu W.J., Lin R.D., Li T.S., Hu Z.H., Ho D.D., Zhang L.Q., Chen Z.W. Natural mutations in the receptor binding domain of spike glycoprotein determine the reactivity of cross-neutralization between palm civet coronavirus and severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:4694–4700. doi: 10.1128/JVI.02389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.E., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- McCray P.B., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., Meyerholz D.K., Kirby P., Look D.C., Perlman S. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W.H., Wong S.K., Li Y.H., Kuhn J.H., Coderre J., Vasilieva N., Han Z.C., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Raj V.S., van Kuppeveld F.J., Rottier P.J., Haagmans B.L., Bosch B.J. The receptor binding domain of the new MERS coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G.Q., Sun D.W., Rajashankar K.R., Qian Z.H., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Nat. Acad. Sci. U.S.A. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G.Q., Xu L.Q., Lin Y.L., Chen L., Pasquarella J.R., Holmes K.V., Li F. Crystal Structure of Bovine Coronavirus Spike Protein Lectin Domain. J. Biol. Chem. 2012;287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Gan J.H., Feng Y., Zhu Z.Y., Choudhry V., Xiao X.D., Ji X.H., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G., Rao X., Song H.D., Wang S.Y., Zuo Y., Zheng A.H., Luo M., Wang H.L., Deng F., Wang H.Z., Hu Z.H., Ding M.X., Zhao G.P., Deng H.K. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H.H., Smits S.L., Dekkers D.H.W., Muller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordoño D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Qu X.X., Li W.D., Han Z.G., Yu M., Zhou P., Zhang S.Y., Wang L.F., Deng H.K., Shi Z.L. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;83:1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., Zaki S.R., Baric R., Subbarao K. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:23–37. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Rockx B., Donaldson E., Corti D., Baric R. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J. Virol. 2008;82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Rockx B., Donaldson E., Sims A., Pickles R., Corti D., Baric R. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82:2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.L., Hu Z.H. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G.D., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S.X., Kong X.G., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Nat. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J.H., Li W.H., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Nat. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T.K., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell. Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W.H., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.L., Chen L., Peng G.Q., Zhou W.B., Pennell C.A., Mansky L.M., Geraghty R.J., Li F. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J. Virol. 2011;85:5331–5337. doi: 10.1128/JVI.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.L., Li W.K., Peng G.Q., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Nat. Acad. Sci. U.S.A. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.L., Peng G.Q., Wilken M., Geraghty R.J., Li F. Mechanisms of Host Receptor Adaptation by Severe Acute Respiratory Syndrome Coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X.D., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil Y., Yagil C. Hypothesis - ACE2 modulates blood pressure in the mammalian organism. Hypertension. 2003;41:871–873. doi: 10.1161/01.HYP.0000063886.71596.C8. [DOI] [PubMed] [Google Scholar]
- Yu I.T.S., Li Y.G., Wong T.W., Tam W., Chan A.T., Lee J.H.W., Leung D.Y.C., Ho T. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 2004;350:1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A., Fouchier R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]


