Abstract
Most emerging infectious diseases (EIDs) in humans have arisen from animals. Identifying high-risk hosts is therefore vital for the control and surveillance of these diseases. Viewing hosts as connected through the parasites they share, we use network tools to investigate predictors of parasitism and sources of future EIDs. We generated host–parasite networks that link hosts when they share a parasite, using nonhuman primates as a model system because—owing to their phylogenetic proximity and ecological overlap with humans—they are an important source of EIDs to humans. We then tested whether centrality in the network of host species—a measurement of the importance of a given node (i.e., host species) in the network—is associated with that host serving as a potential EID source. We found that centrality covaries with key predictors of parasitism, such as population density and geographic range size. Importantly, we also found that primate species having higher values of centrality in the primate–parasite network harbored more parasites identified as EIDs in humans and had parasite communities more similar to those found in humans. These relationships were robust to the use of different centrality metrics and to multiple ways of controlling for variation in how well each species has been studied (i.e., sampling effort). Centrality may therefore estimate the role of a host as a source of EIDs to humans in other multispecific host–parasite networks.
Keywords: ecological networks, epidemiology
Emerging infectious diseases (EIDs) pose a serious threat to human health (1, 2), making the development of tools to predict future EIDs an urgent goal. Most EIDs are zoonotic, that is, they have arisen from animals (3–5), yet our ability to predict sources of new EIDs in humans is remarkably weak, with animal sources of EIDs identified after—rather than in advance of—disease emergence (6). New pandemics might be predicted and prevented if we are able to identify potential EID sources before disease emergence. Efforts are therefore underway to make such predictions on the basis of, for example, phylogenetic relatedness, ecological overlap, and systematic searches for pathogens in wildlife that have higher potential for host shifts (6, 7). Here we show that ecological networks of hosts and parasites predict the occurrence of EIDs in wild primate hosts. Similar approaches can be applied in other host–parasite systems to predict EIDs in humans and wildlife.
Infection dynamics have been investigated extensively using social networks in which nodes represent individuals that are connected through social or sexual contacts (8, 9). Ecological networks involve similar connections among species (rather than individuals) and have been used to investigate a wide range of important questions involving food webs, mutualisms, and antagonistic interactions (10). In host–parasite ecological networks, nodes represent host species that are linked through sharing of parasites (11). Thus, two particular host species are better connected (i.e., they have a stronger edge weight) when they share more parasites in common. Sharing may be generated by several processes: parasites may be shared through new host-shifts (souvenir parasites), through regular pathogen flow of a generalist parasite among hosts, or through common evolutionary descent (heirloom parasites).
Many metrics have been used to investigate the behavior of nodes in networks. One metric, centrality, is especially relevant because it is an indication of the relative importance of the nodes in the network. Centrality can be defined as the connection intensity of a given node with the other nodes in the network (12) (SI Appendix, SI Materials and Methods provides a mathematical definition of centrality). In parasitic networks, host centrality is related to the number of parasites infecting a host (13) and to the parasite’s host range and transmission ability at the level of the whole network (12). Thus, to be central, a host is infected by many parasites that infect many other hosts in the network (14, 15). Consequently, the centrality of a given host could be a good estimate of its potential to be a source of harmful pathogens (15–17). Central individuals are considered in epidemiology as “super-spreaders” because they receive and transmit pathogens more frequently than noncentral individuals (15, 18). Species can also act as superspreaders when they transmit parasites very often to other species. The identification of species that behave as superspreaders is crucial for developing surveillance protocols and interventions aimed at preventing future disease emergence in human populations.
Most primates harbor multiple parasites (19). Primates share a high proportion of their parasite communities (20), with sharing dependent on geographic range overlap, ecological overlap, and phylogeny (7, 20). Parasite sharing may thus occur through common descent or through cross-species transmission. Although humans share more parasites with domesticated animals than with primates (21, 22), some studies suggest that we are more vulnerable to cross-infection from our closest relatives (5, 21). Some of the most harmful diseases in humans have probably transmitted from primates, such as falciparum malaria, yellow fever, and HIV (5, 19).
Results and Discussion
We obtained 6,304 records of parasite and pathogen from the Global Mammal Parasite Database, representing 300 parasite species (including viruses, bacteria, helminths, protozoa, arthropods, and fungi) infecting 140 primate hosts (23). We built a presence/absence bipartite network linking each primate species with their parasites. We then obtained the weighted unipartite projection, in which each node was a primate species that was connected to other primates on the basis of the number of parasite species they shared. Centrality was estimated using five metrics: strength, Opsahl degree, eigenvector, betweenness, and closeness (14, 24). Each of these metrics captures different and complementary aspects of centrality (SI Appendix, SI Materials and Methods). Whereas strength, Opsahl degree, and eigenvector centrality are more related to the pattern of direct cosharing of parasites with the rest of the hosts in the network, betweenness and closeness capture aspects of indirect sharing through other species across the entire network (8, 12, 14, 15).
Centrality may also be affected by the number of studies that have been conducted on each primate species (i.e., sampling effort), because more heavily sampled primates may seem to have more parasites (7, 19). Thus, to get an accurate estimate of centrality requires controlling for variation in sampling effort among primates. We used the number of citations (=number of studies) as an estimate of sampling effort for each primate (25). At present, different methods of controlling for sampling effort are available, and it is unclear which method should be used. To ensure that our findings are robust, we therefore used four different approaches (I–IV) of controlling for variation in sampling effort: I, including sampling effort in the computation of centrality estimates by up-weighting those primates that have more parasites than expected given their sampling effort; II, including sampling effort in the computation of centrality estimates by up-weighting the least sampled primates and down-weighting the most sampled primates; III, obtaining the residuals by regressing centrality on measures of sampling effort (25); and IV, using only the subset of 38 well-sampled primates from ref. 7. A full explanation of the four weighting methods can be found in SI Appendix, SI Materials and Methods. In addition, we ran all analyses in methods I–III including sampling effort as a covariate. Results were consistent across the methods. For the sake of brevity, we provide results from method I in the main text (with other results provided in SI Appendix, SI Results).
All five centrality indices showed positive correlations (0.311 < r < 0.998, P < 0.0001 in all cases, n = 140 primates; SI Appendix, Tables S1, S6, and S13), indicating that they detected similar primate species as most central. Betweenness was the centrality index least correlated with the other indices (0.311 < r < 0.690, P < 0.0001). A main characteristic of betweenness is that, irrespective of the network, many nodes attain a score of zero, resulting in a zero-inflated distribution. This characteristic is thus responsible for the weaker correlation between betweenness and the other centrality metrics. To get a clearer picture of the effect of centrality on primate transmission ability, we obtained a composite index that integrates the different and complementary aspects of the five centrality metrics by performing a principal component analysis on the centrality index correlations. This analysis found a single factor explaining between 77% and 84% of the variance of the indices, depending on the weighting method (SI Appendix, Tables S2, S7, and S14). This factor, hereafter called “centrality,” is used in the subsequent analyses (results for different measures of centrality are provided in SI Appendix, Results). The top 10 most central primates were, in descending order, Papio ursinus, Saimiri sciureus, Alouatta seniculus, Chlorocebus aethiops, Piliocolobus badius, Papio cynocephalus, Papio anubis, Gorilla gorilla, Pan troglodytes, and Macaca sinica (Fig. 1A).
Fig. 1.
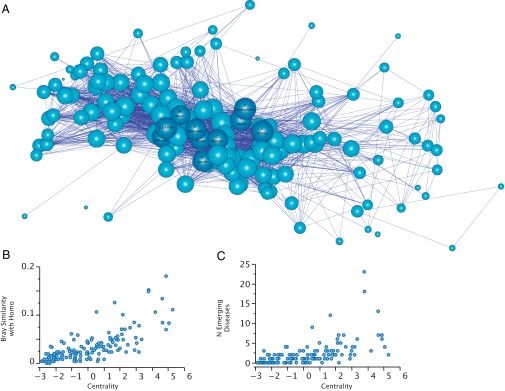
Central primates shared more emerging infectious diseases with humans. (A) Unipartite network depicting the pattern of shared parasites by primates species. Each node represents a primate species. The links among nodes depict shared parasites (i.e., two nodes are linked whenever they share a parasite species). The network representation was generated with the Kamada-Kawai energy-minimization algorithm, which associates the mathematical centrality—calculated as the first factor of a principal component analysis of the five centrality metrics computed for each primate species—with the topological centrality in the network. Thus, nodes in the center of the presented network are more central than nodes in the periphery. The size of the nodes is proportional to the number of EIDs. Dark blue, the top 10 primates sharing more EIDs with humans. (B) Relationship between centrality and the similitude between human and primate parasite communities based on the Bray index. (C) Relationship between centrality and EiD richness found in a primate species.
Several traits have been found to affect parasitism, such as population density, geographic range size, group size, body mass, and diet (19). We explored whether these traits also covary with centrality of the primates using phylogenetic generalized least squares (PGLS) models (26) with the primate consensus dated tree from the 10kTrees Project (27). In a model with multiple predictor variables, we found that primates with denser populations living in larger groups and having broad distributions were more central [population density: estimate ± 1 SE = 0.32 ± 0.15, t = 2.10, P = 0.04; geographic range size: estimate ± 1 SE = 0.28 ± 0.14, t = 1.95, P = 0.05; group size: estimate ± 1 SE = 0.36 ± 0.17, t = 2.08, P = 0.04, R2 = 0.490; PGLS with phylogenetic signal (λ) = 0.138, P = 0.06 compared to a model with λ = 0, n = 122 primates]. This outcome was similar for each of the centrality indices considered independently (SI Appendix, Table S3). We suggest that primates with a large geographic range size and denser populations will have a higher probability of interspecific encounters with other primate species, enhancing the conditions for parasite sharing.
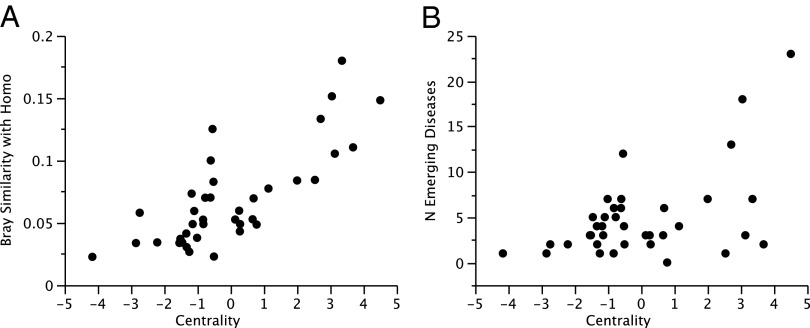
Inferring when, where, and to whom parasites are transmitted are key questions in disease ecology (28). Risk of transmission to humans has been explored by determining the parasites shared both by humans and wild primates (19). Those primate species sharing more pathogens with humans have a higher probability of behaving as zoonotic reservoirs owing to host shifting observed in most primate pathogens (29). To infer the potential ability of different primate species to act as zoonotic reservoirs, we determined the similarity between the parasite communities harbored by primates and humans. Human parasite community and EIDs were obtained from Taylor et al.’s database (30), comprising 1,415 zoonotic and nonzoonotic species. Primates showed variation in the degree to which their parasite community corresponds to that of humans. Some apes, such as G. gorilla (Bray index = 0.16), P. troglodytes (0.15), and Pongo pygmaeus (0.12), had parasite communities similar to those in humans. This outcome is expected owing to the significant phylogenetic conservatism observed in ecological interactions in general (31). Nevertheless, several Old World monkeys also showed similar levels of sharing by this measure, including C. aethiops (0.18), P. cynocephalus (0.13), and P. ursinus (0.11). This finding indicates that in addition to apes, other primates may act as sources of human diseases (7), especially Old World monkeys that show ecological overlap with humans, such as terrestrial substrate use. We found that more central primates had parasite communities that were more similar to humans (estimate ± 1 SE = 0.011 ± 0.002, t = 5.60, P < 0.0001, R2 = 0.750; PGLS with λ = 0.150, P = 0.001, n = 129 primates; Fig. 1B). Again this outcome was consistent across different centrality indices and for most ways of controlling for sampling effort (SI Appendix, Tables S4, S8, and S10). We found the same significant relationship when using only the well-sampled primate species (Fig. 2A and SI Appendix, Table S15).
Fig. 2.
Relationship between network centrality and pathogen infection to humans in well-sampled primates. (A) Relationship between centrality and the similitude between human and primate parasite communities, according to the Bray index. (B) Relationship between centrality and the number of infectious organisms identified as “emerging” in humans (EID richness).
Identifying hosts most frequently associated with EIDs may predict potential pathogen reservoirs and prevent future disease outbreaks (4, 32). We found that, after controlling for sampling effort and phylogeny, the number of infectious organisms found in primates and identified as emerging in humans (EID richness) was significantly related to the centrality of the primates in a multivariate model (Fig. 1C and Table 1). The association with EID richness was significant and positive for each of the five centrality indices, irrespective of the methodology used to control for variation in sampling effort (SI Appendix, Tables S5, S9 , S11, and S16), and maintained when using only the 38 well-sampled primates (Fig. 2B). To assess how phylogenetic uncertainty affects these results, we repeated the analysis using 100 different trees obtained from 10kTrees (27) and found that the association between EID richness and centrality was consistent across all of the phylogenetic trees (SI Appendix, Table S17). In addition, as noted above, centrality is predicted by several ecological and phenotypic traits, such as geographic range size and population density. However, we found that only parasite richness and geographic range size covary with EID richness (Table 1).
Table 1.
Summary of the PGLS multivariate model testing the effects of centrality and several primate traits on EID richness (log-transformed) (n = 122 primate species)
| Parameter | Estimate ± 1 SE | t | P |
| Centrality | 0.252 ± 0.081 | 3.09 | 0.002 |
| Parasite richness | 0.285 ± 0.143 | 1.99 | 0.047 |
| Sampling effort | −0.001 ± 0.112 | 0.02 | 0.980 |
| Body mass (log kg) | 0.006 ± 0.064 | 0.09 | 0.929 |
| Population density (log Ind/km2) | −0.062 ± 0.053 | 1.17 | 0.243 |
| Group size (log Ind) | −0.031 ± 0.060 | 0.51 | 0.612 |
| Geographic range size (log km2) | 0.109 ± 0.051 | 2.13 | 0.035 |
| Diet | |||
| Faunivore vs. folivore | −0.033 ± 0.254 | 0.13 | 0.897 |
| Faunivore vs. frugivore | −0.091 ± 0.241 | 0.38 | 0.707 |
All variables were standardized before analysis. The R2 of the model was 0.526, whereas the phylogenetic signal of the residuals of the model (λ) was estimated to be 0.
Collectively, these analyses suggest that nonhuman primates that are more central in the primate–parasite network also have a higher probability of sharing infectious diseases with humans, including those identified as EIDs. As expected, apes were found among these most-central primates. They are obvious candidates to share infectious organisms with us owing to their phylogenetic proximity, and genetic evidence suggests that spillover has occurred from these species to humans [e.g., falciparum malaria from gorillas (33) and SIV/HIV from chimpanzees (34)]. Nevertheless, we also found that other central non-ape primates, such as baboons and vervet monkeys, are infected by many parasites identified as EIDs in humans.
Our findings suggest that centrality may help to detect risks that might otherwise go unnoticed, and thus to predict disease emergence in advance of outbreaks—an important goal for stemming future zoonotic disease risks (6). For example, with information on the parasites of different host species, effort could focus on sampling individuals of more central species for viruses with higher mutation rates—such as RNA viruses—that have greater potential for spillover to humans and other wildlife. It is important to emphasize that this approach focuses on all parasites and pathogens, regardless of whether they occur in humans; simply having the ecological network is what matters, rather than details on parasites identified as “emerging” or shared with humans. By demonstrating that ecological network characteristics from primates also predict the presence of EIDs in humans, our results validate the use of similar network approaches for predicting future disease emergence in other animal groups that are important sources of EIDs, such as ungulates, rodents, bats, and carnivores.
In conclusion, we provide an approach based on network theory to detect the probability of EID transmission from wildlife to humans. Centrality could therefore become a useful tool for allocating resources to prevent future emerging diseases.
Materials and Methods
Detailed methods, R scripts, and dataset are in SI Appendix, SI Materials and Methods and Dataset S1.
Network Centrality.
We used five centrality metrics: strength and Opsahl degree assess the importance of a node according to its reachability within a network, the former only considering the total number of parasites shared with other primates, and the latter including not only the number of shared parasites but also the number of sharing primates (14, 24). Eigenvector centrality is a variant of degree in which it is assumed that a given node affects all of the neighboring nodes simultaneously (14). Any node is more central under this perspective when it is connected to many highly connected nodes (14). Betweenness is the number of shortest paths between two nodes that pass through a node of interest, and it describes the importance of a node as an intermediary between different parts of the network (12). Closeness is the inverse sum of the shortest distances to all other nodes from a focal node and describes how close a given node is from the rest of the nodes in the network. Betweenness and closeness define the flow pathways and have been shown to be important in the spread of infectious agents (14).
Comparative Analyses.
PGLS (26) methods were used to test the relationship between centrality and (i) primate traits (body mass, geographic range size, population density, group size, parasite richness, and diet), (ii) similarity between primate and human parasite community, and (iii) number of EIDs.
Supplementary Material
Acknowledgments
We thank Randi Griffin, Amy Pedersen, Rosa Menéndez, Mark Lineham, and two anonymous reviewers for discussion and comments on a previous draft. This work was funded by the Spanish Ministry of Science (J.M.G. and M.V.), by the Junta de Andalucía (J.M.G.), and by National Science Foundation Grants DEB-0211908 and EF-0723939/0904359 (C.L.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220716110/-/DCSupplemental.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolhouse MEJ. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 2002;10(10,Suppl):S3–S7. doi: 10.1016/s0966-842x(02)02428-9. [DOI] [PubMed] [Google Scholar]
- 4.Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20(5):238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447(7142):279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morse SS, et al. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380(9857):1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper N, et al. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol Lett. 2012;15(12):1370–1377. doi: 10.1111/j.1461-0248.2012.01858.x. [DOI] [PubMed] [Google Scholar]
- 8.Newman MEJ. Spread of epidemic disease on networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;66(1 Pt 2):016128. doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- 9.Smilkov D, Kocarev L. Influence of the network topology on epidemic spreading. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85(1 Pt 2):016114. doi: 10.1103/PhysRevE.85.016114. [DOI] [PubMed] [Google Scholar]
- 10.Bascompte J, Jordano P. Plant-animal mutualistic networks: the architecture of biodiversity. Annu Rev Ecol Evol Syst. 2007;38:567–593. [Google Scholar]
- 11.Poulin R. Network analysis shining light on parasite ecology and diversity. Trends Parasitol. 2010;26(10):492–498. doi: 10.1016/j.pt.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 13.Anderson TK, Sukhdeo MVK. Host centrality in food web networks determines parasite diversity. PLoS ONE. 2011;6(10):e26798. doi: 10.1371/journal.pone.0026798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opsahl T, Agneessens F, Skvoretz J. Node centrality in weighted networks: generalizing degree and shortest paths. Soc Networks. 2010;32:245–251. [Google Scholar]
- 15.Canright GS, Engoe-Monsen K. Spreading on networks: A topographic view. Complexus. 2006;3:131–146. [Google Scholar]
- 16.Craft ME, Volz E, Packer C, Meyers LA. Distinguishing epidemic waves from disease spillover in a wildlife population. Proc R Soc Lond B Biol Sci. 2009;276(1663):1777–1785. doi: 10.1098/rspb.2008.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordes F, Morand S. The impact of multiple infections on wild animal hosts: A review. Infection Ecol. Epidemiol. 2011;1:7346. doi: 10.3402/iee.v1i0.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin RH, Nunn CL. Community structure and the spread of infectious disease in primate social networks. Evol Ecol. 2012;26:779–800. [Google Scholar]
- 19.Nunn CL, Altizer S. Infectious Diseases in Primates: Behavior, Ecology and Evolution. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 20.Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc R Soc Lond B Biol Sci. 2008;275(1643):1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen AB, Davies TJ. Cross-species pathogen transmission and disease emergence in primates. EcoHealth. 2009;6(4):496–508. doi: 10.1007/s10393-010-0284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss RA. The Leeuwenhoek Lecture 2001. Animal origins of human infectious disease. Philos Trans R Soc Lond B Biol Sci. 2001;356(1410):957–977. doi: 10.1098/rstb.2001.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunn CL, Altizer SM. The Global Mammal Parasite Database: An online resource for infectious disease records in wild primates. Evol Anthropol. 2005;14:1–2. [Google Scholar]
- 24.Barrat A, Barthélemy M, Pastor-Satorras R, Vespignani A. The architecture of complex weighted networks. Proc Natl Acad Sci USA. 2004;101(11):3747–3752. doi: 10.1073/pnas.0400087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunn CL, Altizer S, Jones KE, Sechrest W. Comparative tests of parasite species richness in primates. Am Nat. 2003;162(5):597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
- 26.Nunn CL. The Comparative Approach in Evolutionary Anthropology and Biology. Chicago: Univ of Chicago Press; 2011. [Google Scholar]
- 27.Arnold C, Matthews LJ, Nunn CL. The 10kTrees website: A new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- 28.Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: A new frontier in disease ecology. Trends Ecol Evol. 2009;24(1):21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Garamszegi LZ. Patterns of co-speciation and host switching in primate malaria parasites. Malar J. 2009;8:110. doi: 10.1186/1475-2875-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356(1411):983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gómez JM, Verdú M, Perfectti F. Ecological interactions are evolutionarily conserved across the entire tree of life. Nature. 2010;465(7300):918–921. doi: 10.1038/nature09113. [DOI] [PubMed] [Google Scholar]
- 32.Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. Identifying reservoirs of infection: A conceptual and practical challenge. Emerg Infect Dis. 2002;8(12):1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287(5453):607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.